Abstract
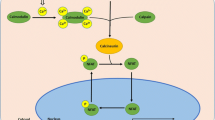
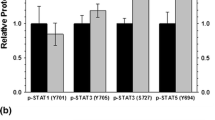
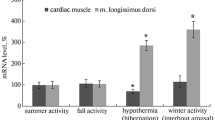
The thirteen-lined ground squirrel (Ictidomys tridecemlineatus) undergoes remarkable adaptive changes during hibernation. Interestingly, skeletal muscle remodelling occurs during the torpor-arousal cycle of hibernation to prevent net muscle loss despite inactivity. Reversible cardiomyocyte hypertrophy occurs in cardiac muscle, allowing the heart to preserve cardiac output during hibernation, while avoiding chronic maladaptive hypertrophy post-hibernation. We propose that calcium signalling proteins [calcineurin (Cn), calmodulin (CaM), and calpain], the nuclear factor of activated T cell (NFAT) family of transcription factors, and the NFAT targets myoferlin and myomaker contribute significantly to adaptations taking place in skeletal and cardiac muscle during hibernation. Protein-level analyses were performed over several conditions: euthermic room temperature (ER), euthermic cold room (EC), entrance into (EN), early (ET), and late torpor (LT) time points, in addition to early (EA), interbout (IA), and late arousal (LA) time points using immunoblotting and DNA–protein interaction (DPI) enzyme-linked immunosorbent assay (ELISAs). In skeletal and cardiac muscle, NFATc2 protein levels were elevated during torpor. NFATc4 increased throughout the torpor-arousal cycle in both tissues, and NFATc1 showed this trend in cardiac muscle only. NFATc3 showed an elevation in DNA-binding activity but not expression during torpor. Myoferlin protein levels dramatically increased during torpor in both skeletal and cardiac muscle. Myomaker levels also increased significantly in cardiac muscle during torpor. Cardiac Cn levels remained stable, whereas CaM and calpain decreased throughout the torpor-arousal cycle. Activation and/or upregulation of NFATc2, c3, myoferlin, and myomaker at torpor could be part of a stress-response mechanism to preserve skeletal muscle mass, whereas CaM and calpain appear to initiate the rapid reversal of cardiac hypertrophy during arousal through downregulation of the NFAT–Cn pathway.






Similar content being viewed by others
References
Frerichs KU, Hallenbeck JM (1998) Hibernation in ground squirrels induces state and species-specific tolerance to hypoxia and aglycemia: an in vitro study in hippocampal slices. J Cereb Blood Flow Metab 18:168–175. doi:10.1097/00004647-199802000-00007
Storey KB (2010) Out cold: biochemical regulation of mammalian hibernation—a mini-review. Gerontology 56:220–230. doi:10.1159/000228829
Storey KB, Storey JM (2004) Metabolic rate depression in animals: transcriptional and translational controls. Biol Rev Camb Philos Soc 79:207–233. doi:10.1017/S1464793103006195
Wang LCH, Lee TF (1996) Torpor and hibernation in mammals: metabolic, physiological, and biochemical adaptations. In: Fregley MJ, Blatteis CM (eds) Handbook of physiology: environmental physiology, section 4, vol 1. Oxford University Press, New York, pp 507–532
Pier M, Ni Z, McMullen DC, Storey KB (2008) Expression of Nrf2 and its downstream gene targets in hibernating 13-lined ground squirrels, Spermophilus tridecemlineatus. Mol Cell Biochem 312:121–129. doi:10.1007/s11010-008-9727-3
Fahlman A, Storey JM, Storey KB (2000) Gene up-regulation in heart during mammalian hibernation. Cryobiology 40:332–342. doi:10.1006/cryo.2000.2254
Morin PJ, Storey KB (2007) Antioxidant defense in hibernation: cloning and expression of peroxiredoxins from hibernating ground squirrels, Spermophilus tridecemlineatus. Arch Biochem Biophys 461:59–65. doi:10.1016/j.abb.2007.01.035
Morin P, Storey KB (2006) Evidence for a reduced transcriptional state during hibernation in ground squirrels. Cryobiology 53:310–318. doi:10.1016/j.cryobiol.2006.08.002
Mamady H, Storey KB (2006) Up-regulation of the endoplasmic reticulum molecular chaperone GRP78 during hibernation in thirteen-lined ground squirrels. Mol Cell Biochem 292:89–98. doi:10.1007/s11010-006-9221-8
Malatesta M, Perdoni F, Battistelli S, Muller S, Zancanaro C (2009) The cell nuclei of skeletal muscle cells are transcriptionally active in hibernating edible dormice. BMC Cell Biol 10:19. doi:10.1186/1471-2121-10-19
Choi H, Selpides P-JI, Nowell MM, Rourke BC (2009) Functional overload in ground squirrel plantaris muscle fails to induce myosin isoform shifts. Am J Physiol Regul Integr Comp Physiol 297:R578–R586. doi:10.1152/ajpregu.00236.2009
Bassel-Duby R, Olson EN (2006) Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem 75:19–37. doi:10.1146/annurev.biochem.75.103004.142622
Rourke BC, Yokoyama Y, Milsom WK, Caiozzo VJ (2004) Myosin isoform expression and MAFbx mRNA levels in hibernating golden- mantled ground squirrels (Spermophilus lateralis). Physiol Biochem 77:582–593
Cotton CJ, Harlow HJ (2015) Avoidance of skeletal muscle atrophy in spontaneous and facultative hibernators. Physiol Biochem Zool 83:551–560. doi:10.1086/650471
Gao YF, Wang J, Wang HP, Feng B, Dang K, Wang Q, Hinghofer-Szalkay HG (2012) Skeletal muscle is protected from disuse in hibernating dauria ground squirrels. Comp Biochem Physiol Part A 161:296–300. doi:10.1016/j.cbpa.2011.11.009
Xu R, Andres-Mateos E, Mejias R, MacDonald EM, Leinwand L, Merriman DK, Fink RHA, Cohn RD (2013) Hibernating squirrel muscle activates the endurance exercise pathway despite prolonged immobilization. Exp Neurol 247:392–401. doi:10.1016/j.expneurol.2013.01.005
Frerichs KU, Kennedy C, Sokoloff L, Hallenbeck JM (1994) Local cerebral blood flow during hibernation, a model of natural tolerance to “cerebral ischemia”. J Cereb Blood Flow Metab 14:193–205. doi:10.1038/jcbfm.1994.26
Nakipova OV, Zakharova NM, Andreeva LA, Chumaeva NA, Averin A, Kosarskii LS, Anufriev AI, von Lewinski D, Kockskamper J, Pieske B (2007) The seasonal peculiarities of force-frequency relationships in active ground squirrel Spermophilus undulatus ventricle. Cryobiology 55:173–181. doi:10.1016/j.cryobiol.2007.07.001
Wickler SJ, Hoyt DF, van Breukelen F (1991) Disuse atrophy in the hibernating golden-mantled ground squirrel, Spermophilus lateralis. Am J Physiol 261:R1214–R1217
Frey N, Katus HA, Olson EN, Hill JA (2004) Hypertrophy of the heart: a new therapeutic target? Circulation 109:1580–1589. doi:10.1161/01.CIR.0000120390.68287.BB
Li H, Liu T, Chen W, Jain MR, Vatner DE, Vatner SF, Kudej RK, Yan L (2013) Proteomic mechanisms of cardioprotection during Mammalian hibernation in woodchucks, Marmota Monax. J Proteome Res 12:4221–4229. doi:10.1021/pr400580f
Nelson OL, Rourke BC (2013) Increase in cardiac myosin heavy-chain (MyHC) alpha protein isoform in hibernating ground squirrels, with echocardiographic visualization of ventricular wall hypertrophy and prolonged contraction. J Exp Biol 216:4678–4690. doi:10.1242/jeb.088773
Tessier SN, Storey KB (2012) Myocyte enhancer factor-2 and cardiac muscle gene expression during hibernation in thirteen-lined ground squirrels. Gene 501:8–16. doi:10.1016/j.gene.2012.04.004
Yan L, Kudej RK, Vatner DE, Vatner SF (2015) Myocardial ischemic protection in natural mammalian hibernation. Basic Res Cardiol. doi:10.1007/s00395-015-0462-0
Rao A, Luo C, Hogan PG (1997) Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 15:707–747. doi:10.1146/annurev.immunol.15.1.707
Rusnak F, Mertz P (2000) Calcineurin: form and function. Physiol Rev 80:1483–1521. doi:10.1172/JCI57909.date
Shibasaki F, Hallin U, Uchino H (2002) Calcineurin as a multifunctional regulator. J Biochem 131:1–15
Yang S, Klee CB (2000) Low affinity Ca2+ -binding sites of calcineurin B mediate conformational changes in calcineurin A. Biochemistry 39:16147–16154
Al-Shanti N, Stewart CE (2009) Ca2+/calmodulin-dependent transcriptional pathways: potential mediators of skeletal muscle growth and development. Biol Rev 84:637–652. doi:10.1111/j.1469-185X.2009.00090.x
Burkard N (2005) Targeted proteolysis sustains calcineurin activation. Circulation 111:1045–1053. doi:10.1161/01.CIR.0000156458.80515.F7
Wu HY, Tomizawa K, Oda Y, Wei FY, Lu YF, Matsushita M, Li ST, Moriwaki A, Matsui H (2004) Critical role of calpain-mediated cleavage of calcineurin in excitotoxic neurodegeneration. J Biol Chem 279:4929–4940. doi:10.1074/jbc.M309767200
Lee SH, Choi J, Kim H, Lee DH, Roh GS, Kim HJ, Kang SS, Choi WS, Cho GJ (2014) FK506 reduces calpain-regulated calcineurin activity in both the cytoplasm and the nucleus. Anat Cell Biol 47:91–100. doi:10.5115/acb.2014.47.2.91
Shioda N, Moriguchi S, Shirasaki Y, Fukunaga K (2006) Generation of constitutively active calcineurin by calpain contributes to delayed neuronal death following mouse brain ischemia. J Neurochem 98:310–320. doi:10.1111/j.1471-4159.2006.03874.x
Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li P-F (2009) miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci USA 106:12103–12108. doi:10.1073/pnas.0811371106
Liu Q, Wilkins BJ, Lee YJ, Ichijo H, Molkentin JD (2006) Direct interaction and reciprocal regulation between ASK1 and calcineurin–NFAT control cardiomyocyte death and growth. Mol Cell Biol 26:3785–3797. doi:10.1128/MCB.26.10.3785
Schubert W, Yang XY, Yang TTC, Factor SM, Lisanti MP, Molkentin JD, Rincon M, Chow C-W (2003) Requirement of transcription factor NFAT in developing atrial myocardium. J Cell Biol 161:861–874. doi:10.1083/jcb.200301058
Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN (1998) A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 93:215–228. doi:10.1016/S0092-8674(00)81573-1
Armand A-S, Bourajjaj M, Martínez-Martínez S, el Azzouzi H, da Costa Martins PA, Hatzis P, Seidler T, Redondo JM, De Windt LJ (2008) Cooperative synergy between NFAT and MyoD regulates myogenin expression and myogenesis. J Biol Chem 283:29004–29010. doi:10.1074/jbc.M801297200
Delling U, Tureckova J, Lim HAEW, Windt LJDE, Rotwein P, Molkentin JD (2000) A Calcineurin–NFATc3-dependent pathway regulates skeletal muscle differentiation and slow myosin heavy-chain expression. Mol Cell Biol 20:6600–6611
Hudson MB, Woodworth-Hobbs ME, Zheng B, Rahnert J, Blount MA, Gooch JL, Searles CD, Price SR (2014) miR-23a is decreased during muscle atrophy by a mechanism that includes calcineurin signaling and exosome-mediated export. Am J Physiol Cell Physiol 306:C551–C558. doi:10.1152/ajpcell.00266.2013
Schiaffino S, Sandri M, Murgia M (2007) Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda) 22:269–278. doi:10.1152/physiol.00009.2007
Tessier SN, Storey KB (2010) Expression of myocyte enhancer factor-2 and downstream genes in ground squirrel skeletal muscle during hibernation. Mol Cell Biochem 344:151–162. doi:10.1007/s11010-010-0538-y
Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK (2001) Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol 153:329–338. doi:10.1083/jcb.153.2.329
Davis DB, Delmonte AJ, Ly CT, McNally EM (2000) Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum Mol Genet 9:217–226
Davis DB, Doherty KR, Delmonte AJ, McNally EM (2002) Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains. J Biol Chem 277:22883–22888. doi:10.1074/jbc.M201858200
Doherty KR, Cave A, Davis DB, Delmonte AJ, Posey A, Earley JU, Hadhazy M, McNally EM (2005) Normal myoblast fusion requires myoferlin. Development 132:5565–5575. doi:10.1242/dev.02155
Haslett JN, Sanoudou D, Kho AT, Han M, Bennett RR, Kohane IS, Beggs AH, Kunkel LM (2003) Gene expression profiling of Duchenne muscular dystrophy skeletal muscle. Neurogenetics 4:163–171. doi:10.1007/s10048-003-0148-x
Millay DP, O’Rourke JR, Sutherland LB, Bezprozvannaya S, Shelton JM, Bassel-Duby R, Olson EN (2013) Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499:301–305. doi:10.1038/nature12343
Millay DP, Sutherland LB, Bassel-duby R, Olson EN (2014) Myomaker is essential for muscle regeneration Myomaker is essential for muscle regeneration. Genes Dev 28(15):1641–1646. doi:10.1101/gad.247205.114
Demonbreun AR, Lapidos KA, Heretis K, Levin S, Dale R, Pytel P, Svensson EC, McNally EM (2010) Myoferlin regulation by NFAT in muscle injury, regeneration and repair. J Cell Sci 123:2413–2422. doi:10.1242/jcs.065375
McMullen DC, Hallenbeck JM (2010) Regulation of Akt during torpor in the hibernating ground squirrel, Ictidomys tridecemlineatus. J Comp Physiol B 180:927–934. doi:10.1007/s00360-010-0468-8
Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38:695–699. doi:10.1093/nar/gkq313
McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, Cowley AP, Lopez R (2013) Analysis tool web services from the EMBL-EBI. Nucleic Acids Res 41:597–600. doi:10.1093/nar/gkt376
Eaton SL, Roche SL, Llavero Hurtado M, Oldknow KJ, Farquharson C, Gillingwater TH, Wishart TM (2013) Total protein analysis as a reliable loading control for quantitative fluorescent western blotting. PLoS One 8:1–9. doi:10.1371/journal.pone.0072457
Hung H-F, Wang B-W, Chang H, Shyu K-G (2008) The molecular regulation of resistin expression in cultured vascular smooth muscle cells under hypoxia. J Hypertens 26:2349–2360. doi:10.1097/HJH.0b013e328311fa30
Brand LH, Kirchler T, Hummel S, Chaban C, Wanke D (2010) DPI–ELISA: a fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods 6:25. doi:10.1186/1746-4811-6-25
Brand LH, Henneges C, Schüssler A, Kolukisaoglu HÃ, Koch G, Wallmeroth N, Hecker A, Thurow K, Zell A, Harter K, Wanke D (2013) Screening for protein-DNA interactions by automatable DNA-protein interaction ELISA. PLoS One 8:e75177. doi:10.1371/journal.pone.0075177
Jagelska E, Brázda V, Pospisilová S, Vojtesek B, Palecek E (2002) New ELISA technique for analysis of p53 protein/DNA binding properties. J Immunol Methods 267:227–235. doi:10.1016/S0022-1759(02)00182-5
Hindle AG, Otis JP, Epperson LE, Hornberger TA, Goodman CA, Carey HV, Martin SL (2014) Prioritization of skeletal muscle growth for emergence from hibernation. J Exp Biol 218:276–284. doi:10.1242/jeb.109512
Cho YY, Yao K, Bode AM, Bergen HR, Madden BJ, Oh SM, Ermakova S, Bong SK, Hong SC, Shim JH, Dong Z (2007) RSK2 mediates muscle cell differentiation through regulation of NFAT3. J Biol Chem 282:8380–8392. doi:10.1074/jbc.M611322200
Wu CW, Storey KB (2014) FoxO3a-mediated activation of stress responsive genes during early torpor in a mammalian hibernator. Mol Cell Biochem 1–11. doi:10.1007/s11010-014-1969-7
Foletta VC, White LJ, Larsen AE, Léger B, Russell AP (2011) The role and regulation of MAFbx/atrogin-1 and MuRF1 in skeletal muscle atrophy. Pflug Arch 461:325–335. doi:10.1007/s00424-010-0919-9
Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M (2013) Mechanisms regulating skeletal muscle growth and atrophy. FEBS J 280:4294–4314. doi:10.1111/febs.12253
Day SM (2013) The ubiquitin proteasome system in human cardiomyopathies and heart failure. Am J Physiol Heart Circ Physiol 304:H1283–H1293. doi:10.1152/ajpheart.00249.2012
de Andrade LHS, de Moraes WMAM, Matsuo Junior EH, de Orleans Carvalho de Moura E, Antunes HKM, Montemor J, Antonio EL, Bocalini DS, Serra AJ, Tucci PJE, Brum PC, Medeiros A (2015) Aerobic exercise training improves oxidative stress and ubiquitin proteasome system activity in heart of spontaneously hypertensive rats. Mol Cell Biochem 402:193–202. doi:10.1007/s11010-015-2326-1
Acknowledgments
We thank Dr. J. M. Hallenbeck and Dr. D.C. McMullen (NINDS, NIH, Bethesda) for providing the tissue samples for this study. Thanks also to J. M. Storey for editorial review of the manuscript. This work was supported by a Discovery grant from the Natural Sciences and Engineering Research Council (NSERC) of Canada (# 6793) and a grant from the Heart and Stroke Foundation of Canada (#G-14-0005874) to KBS. KBS holds the Canada Research Chair in Molecular Physiology; YZ held a postgraduate Queen Elizabeth II Graduate Scholarship in Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Storey, K.B. Expression of nuclear factor of activated T cells (NFAT) and downstream muscle-specific proteins in ground squirrel skeletal and heart muscle during hibernation. Mol Cell Biochem 412, 27–40 (2016). https://doi.org/10.1007/s11010-015-2605-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-015-2605-x