Abstract
Context
The natural patchiness of wetlands and flooding events are likely to strongly affect the genetic structure of their terrestrial species. However, these effects are not well understood yet, especially for small mammals.
Objectives
We investigated at different spatial scales the genetic structure of the harvest mouse (Micromys minutus), a threatened small mammal strongly tied to wetlands, and the effects on gene flow of flooding and of the different types of landscape elements composing a wetland.
Methods
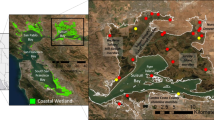
309 harvest mice were sampled in eight sites in Western Europe. Their genetic structure and diversity at 15 microsatellite loci were analyzed at a regional spatial scale and at a local scale, where the resistance of land cover and flooding to gene flow was also assessed, with the optimization procedure implemented in ResistanceGA.
Results
At a regional scale, our study revealed a strong genetic differentiation between populations from Northern Europe to the Mediterranean Sea. At the local scale, in a flooded wetland in France, the species exhibited a large genetic cluster over at least 45 km2, in spite of a large river crossing it. Winter floods explained genetic structure better than landscape features alone, with a stronger resistance to gene flow in reed beds where vegetation level above water was high: contrary to meadows, from which individuals are forced to escape, reed beds can be “golden prisons”, i.e. refuges during floods but with very low possibility of movements.
Conclusions
Several parameters influencing the functioning of a flooded population of harvest mouse are here highlighted that can also be useful for the development of plans to safeguard wetland ecosystems.






Similar content being viewed by others
References
Aplin K, Lunde D, Batsaikhan N, Kryštufek B, Meinig H, Henttonen H (2016) Micromys minutus. IUCN Red List of Threatened Species. http://www.iucnredlist.org. Accessed 04.01.2019.
Amori G, Contoli L, Nappi A (2008) Fauna d’Italia. Mammalia II, Calderini, Bologna
Balčiauskas L, Balčiauskienė L, Janonytė A (2012) The influence of spring floods on small mammal communities in the Nemunas River Delta, Lithuania. Biologia (Bratisl) 67:1220–1229.
Berckmoes V, Scheirs J, Jordaens K, Blust R, Backeljau T, Verhagen R (2005) Effects of environmental pollution on microsatellite DNA diversity in Wood mouse (Apodemus sylvaticus) populations. Environ Toxicol Chem 24:2898.
Berthier K, Galan M, Foltête JC, Charbonnel N, Cosson JF (2005) Genetic structure of the cyclic fossorial Water vole (Arvicola terrestris): landscape and demographic influences. Mol Ecol 14:2861–2871.
Blant M, Marchesi P, Descombes M, Capt S (2012) Nouvelles données sur la répartition de la souris des moissons (Micromys minutus Pallas, 1771) en Suisse occidentale et implications pour la gestion de son habitat. Rev Suisse Zool 119:485–500
Booth W, Montgomery WI, Prodöhl PA (2009) Spatial genetic structuring in a vagile species, the European wood mouse. J Zool 279:219–228.
Churchfield S, Hollier J, Brown VK (1997) Community structure and habitat use of small mammals in grasslands of different successional age. J Zool 242:519–530
Clarke RT, Rothery P, Raybould AF (2002) Confidence limits for regression relationships between distance matrices: estimating gene flow with distance. J Agric Biol Environ Stat 7:361–372
Coulon A, Fitzpatrick JW, Bowman J, Stith BM, Makarewich CA, Stenzler LM, Lovette IJ (2008) Congruent population structure inferred from dispersal behavior and intensive genetic surveys of the threatened Florida Scrub-Jay (Aphelocoma coerulescens). Mol Ecol 17:1685–1701
Cunnings A, Johnson E, Martin Y (2016) Fluvial seed dispersal of riparian trees: transport and depositional processes. Earth Surf Process Landf 41:615–625.
Darinot F (2016) The Harvest mouse (Micromys minutus Pallas, 1771) as prey: a literature review. Folia Zool 65:117–137.
Darinot F (2019) Dispersion et structure génétique d’une population de Rat des moissons (Micromys minutus PALLAS, 1771) soumise à des inondations régulières. EPHE PSL research University Ph.D. thesis.
Després L, Henniaux C, Rioux D, Capblancq T, Zupan S, Čelik T, Sielezniew M, Bonato L, Ficetola GF (2019) Inferring the biogeography and demographic history of an endangered butterfly in Europe from multilocus markers. Biol J Linn Soc 126:95–113
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361.
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620.
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. 22
Finlayson CM, D’Cruz R, Davidson N (2005) Ecosystems and human well-being: wetlands and water: synthesis. World Resources Institute, Washington, DC
Fraaije RGA, Moinier S, van Gogh I, Timmers R, van Deelen JJ, Verhoeven JTA, Soons MB (2017) Spatial patterns of water-dispersed seed deposition along stream riparian gradients. PLoS ONE 12:e0185247.
Frank F (1957) Zucht und Gefangenschafts-biologie der Zwergmaus (Micromys minutus subobscurus, Fritsche). Z Sáugetierk 22:1–44
Garrido-Garduño T, Téllez-Valdés O, Manel S, Vázquez-Domínguez E (2016) Role of habitat heterogeneity and landscape connectivity in shaping gene flow and spatial population structure of a dominant rodent species in a tropical dry forest. J Zool 298:293–302.
Gauffre B, Estoup A, Bretagnolle V, Cosson JF (2008) Spatial genetic structure of a small rodent in a heterogeneous landscape. Mol Ecol 17:4619–4629.
Gibbs JP (1993) Importance of small wetlands for the persistence of local populations of wetland-associated animals. Wetlands 13:25–31.
Goudet J (2004) HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186.
Harris S (1979) Breeding season, litter size and nestling mortality of the Harvest mouse, Micromys minutus (Rodentia: Muridae), in Britain. J Zool 188:437–442.
Hartmann SA, Steyer K, Kraus RHS, Segelbacher G, Nowak C (2013) Potential barriers to gene flow in the endangered European wildcat (Felis silvestris). Conserv Genet 14:413–426.
Heckel G, Burri R, Fink S, Desmet JF, Excoffier L (2005) Genetic structure and colonization processes in European populations of the Common vole, Microtus arvalis. Evolution 59:2231–2242.
Hesselbarth MHK, Sciaini M, With KA, Wiegand K, Nowosad J (2019) Landscapemetrics: an open-source R tool to calculate landscape metrics. Ecography 42:1648–1657.
Jaarola M, Searle JB (2002) Phylogeography of Field voles (Microtus agrestis) in Eurasia inferred from mitochondrial DNA sequences: phylogeography of Field voles. Mol Ecol 11:2613–2621.
Janes JK, Miller JM, Dupuis JR, Malenfant RM, Gorrell JC, Cullingham CI, Andrew RL (2017) The K=2 conundrum. Mol Ecol 26:3594–3602.
Jiang S-Y, Lin YK (2009) Polymorphic Microsatellite Markers for the Harvest Mouse (Micromys minutus) in Taiwan. Taiwania 54:118–121
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405.
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94.
Kalinowski ST (2005) hp-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189.
Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35:43–46.
Klee RV, Mahoney AC, Christopher CC, Barrett GW (2004) Riverine peninsulas: an experimental approach to homing in White-footed Mice (Peromyscus leucopus). Am Midl Nat 151:408–413
Kittlein MJ, Gaggiotti OE (2008) Interactions between environmental factors can hide isolation by distance patterns: a case study of Ctenomys rionegrensis in Uruguay. Proc R Soc B 275:2633–2638.
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191.
Lugon-Moulin N, Nner HB, Hausser J, Goudet JRM (1999) Do riverine barriers, history or introgression shape the genetic structuring of a Common shrew (Sorex araneus) population? Heredity 83:155–161
Malausa T, Gilles A, Meglécz E, Blanquart H, Duthoy S, Costedoat C, Dubut V, Pech N, Castagnone-Sereno P, Délye C, Feau N, Frey P, Gauthier P, Guillemaud T, Hazard L, Le Corre V, Lung-Escarmant B, Malé PJG, Ferreira S, Martin JF (2011) High-throughput microsatellite isolation through 454 GS-FLX titanium pyrosequencing of enriched DNA libraries: pyrosequencing of SSR-enriched DNA libraries. Mol Ecol Resour 11:638–644
Matocq MD, Patton JL, da Silva MNF (2000) Population genetic structure of two ecologically distinct Amazonian spiny rats: separating history and current ecology. Evolution 54:1423–1432
McRae BH, Beier P (2007) Circuit theory predicts gene flow in plant and animal populations. Proc Natl Acad Sci 104:19885–19890
McRae BH, Dickson BG, Keitt TH, Shah VB (2008) Using circuit theory to model connectivity in ecology, evolution, and conservation. Ecology 89:2712–2724
Meirmans PG (2012) The trouble with isolation by distance. Mol Ecol 21:2839–2846
Mergey M, Bardonnet C, Quintaine T (2017) Identifying environmental drivers of spatial genetic structure of the European pine marten (Martes martes). Landsc Ecol 32:2261–2279.
Patton JL, da Silva MNF, Malcolm JR (1994) Gene genealogy and differentiation among arboreal spiny rats (Rodentia: Echimyidae) of the Amazon basin: a test of the riverine barrier hypothesis. Evolution 48:1314–1323.
Peakall R, Smouse PE (2006) genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295.
Pereoglou F, Lindenmayer DB, MacGregor C, Ford F, Wood J, Banks SC (2013) Landscape genetics of an early successional specialist in a disturbance-prone environment. Mol Ecol 22:1267–1281.
Perez MF, Franco FF, Bombonato JR, Bonatelli IAS, Khan G, Romeiro-Brito M, Fegies AC, Ribeiro PM, Silva GAR, Moraes EM (2018) Assessing population structure in the face of isolation by distance: are we neglectingthe problem? Divers Distrib 24:1883–1889
Perrow M, Jowitt A (1995) What future for the Harvest mouse? British Wildlife 6:356–365
Peterman WE (2018) ResistanceGA: an R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol Evol 9:1638–1647.
Peterman WE, Connette GM, Semlitsch RD, Eggert LS (2014) Ecological resistance surfaces predict fine-scale genetic differentiation in a terrestrial woodland salamander. Mol Ecol 23:2402–2413.
Porras-Hurtado L, Ruiz Y, Santos C, Phillips C, Carracedo A, Lareu MV (2013) An overview of STRUCTURE: applications, parameter settings, and supporting software. Front Genet. https://doi.org/10.3389/fgene.2013.00098
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.
Rocha RG, Ferreira E, Fonseca C, Justino J, Leite YLR, Pires Costa L (2014) Seasonal flooding regime and ecological traits influence genetic structure of two small rodents. Ecol Evol 4(24):4598–4608
Rousset F (2008) genepop’007: a complete re-implementation of the genepop software for Windows and Linux. Mol Ecol Resour 8:103–106.
Russo IRM, Sole CL, Barbato M, von Bramann U, Bruford MW (2016) Landscape determinants of fine-scale genetic structure of a small rodent in a heterogeneous landscape (Hluhluwe-iMfolozi Park, South Africa). Sci Rep 6:29168
Schooley RL, Branch LC (2009) Enhancing the area–isolation paradigm: habitat heterogeneity and metapopulation dynamics of a rare wetland mammal. Ecol Appl 19:1708–1722.
Schooley RL, Cosentino BJ (2018) Metapopulation dynamics of wetland species. In: Finlayson CM, Everard M, Irvine K et al (eds) The Wetland book. Springer, Netherlands, Dordrecht, pp 141–147
Sheppe W, Haas P (1981) The annual cycle of small mammal populations along the Chobe River, Botswana. Mammalia 45:157–176.
Smith KR, Barthman-Thompson L, Gould WR, Mabry KE (2014) Effects of natural and anthropogenic change on habitat use and movement of endangered Salt Marsh Harvest Mice. PLoS ONE 9:e108739.
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538.
Weir BS, Cockerham CC (1984) Estimating F -statistics for the analysis of population structure. Evolution 38:1358–1370.
Wijnhoven S, Van der Velde G, Leuven RSEW, Smits AJM (2005) Flooding ecology of voles, mice and shrews: the importance of geomorphological and vegetational heterogeneity in river floodplains. Acta Theriol 50:453–472
Wyttenbach A, Narain Y, Fredga K (1999) Genetic structuring and gene flow in a hybrid zone between two chromosome races of the Common shrew (Sorex araneus, Insectivora) revealed by microsatellites. Heredity 82:79–88.
Zedler JB, Kercher S (2005) Wetland resources: status, trends, ecosystem services, and restorability. Annu Rev Environ Resour 30:39–74.
Zhang M, Wang K, Wang Y, Guo C, Li B, Huang H (2007) Recovery of a rodent community in an agro-ecosystem after flooding. J Zool 272:138–147.
Acknowledgements
The authors would like to thank for people who help them in obtaining tissues of M. minutus for DNA isolation, for the description of new polymorphic microsatellite markers and genotyping of populations: Grégoire Massez and Sylvain Ceyte (Réserve Naturelle Nationale des Marais du Vigueirat), Gérard Hommay (Institut National de la Recherche Agronomique), François Léger (Office National de la Chasse et de la Faune Sauvage), Andrea Schüster and Katarina Foerster (University of Tübingen), Jacques Gilliéron and Hubert du Plessix (Prodon marsh), Maurice Benmergui (Office National de la Chasse et de la Faune Sauvage, Birieux pond), Olivier Glaizot (Cantonal Museum of Zoology of Lausanne, Neuchâtel lake), Emilie Wichroff and Rémi Bogey (Syndicat du Haut-Rhône, Saugey meander), Manuel Bouron (Conservatoire des Espaces Naturels de la Savoie, Chautagne marsh). This study was supported by the French Ministry of Environment and the Departmental Council of Ain.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Darinot, F., Le Petitcorps, Q., Arnal, V. et al. Effects of landscape features and flooding on the genetic structure of a small wetland rodent, the harvest mouse (Micromys minutus). Landscape Ecol 36, 1755–1771 (2021). https://doi.org/10.1007/s10980-021-01235-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10980-021-01235-5