Abstract
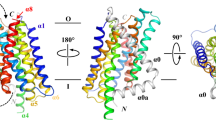
The ZIP family transport zinc ions from the extracellular medium across the plasma membrane or from the intracellular compartments across endomembranes, which play fundamental roles in metal homeostasis and are broadly involved in physiological and pathological processes. Desulfovibrio is the predominant sulphate-reducing bacteria in human colonic microbiota, but also a potential choice for metal bioremediation. while, there are no published studies describing the zinc transporters from Desulfovibrio up to now. In this study, we obtained for the first time a heterologously expressed ZIP homolog from Desulfovibrio vulgaris, termed dvZip. The purified dvZip was reconstituted into proteoliposomes, and confirmed its zinc transport ability in vitro. By combining topology prediction, homology modeling and phylogenetic approaches, we also noticed that dvZip belongs to the GufA and probably have 8 transmembrane α-helical segments (TM 1–8) in which both termini are located on the extracellular, with TM2, 4, 5 and 7 create an inner bundle. We believe that purification and characterization of zinc (probably also cadmium) transporters from Desulfovibrio vulgaris such as dvZip could shed light on understanding of metal homeostasis of Desulfovibrio and provided protein products for future detailed function and structural studies.





Similar content being viewed by others
Data Availability
The PDB file of dvZip model is available from the corresponding author on reasonable request. All the remaining data are included in this article and its supplementary information files.
References
de Luis DA, Izaola O, Aller R, Armentia A, Cuellar L (2003) Antioxidant and fat intake in patients with polinic asthma. Med Clin 121:653–654
Ho E, Quan N, Tsai YH, Lai W, Bray TM (2001) Dietary zinc supplementation inhibits NFkappaB activation and protects against chemically induced diabetes in CD1 mice. Exp Biol Med (Maywood) 226:103–111. https://doi.org/10.1177/153537020122600207
Lang C, Murgia C, Leong M, Tan LW, Perozzi G, Knight D, Ruffin R, Zalewski P (2007) Anti-inflammatory effects of zinc and alterations in zinc transporter mRNA in mouse models of allergic inflammation. Am J Physiol Lung Cell Mol Physiol 292:L577-584. https://doi.org/10.1152/ajplung.00280.2006
Sekler I, Sensi SL, Hershfinkel M, Silverman WF (2007) Mechanism and regulation of cellular zinc transport. Mol Med 13:337–343. https://doi.org/10.2119/2007-00037.Sekler
Soutar A, Seaton A, Brown K (1997) Bronchial reactivity and dietary antioxidants. Thorax 52:166–170. https://doi.org/10.1136/thx.52.2.166
Terres-Martos C, Navarro-Alarcon M, Martin-Lagos F, LopezPerez-Valero GdlSHV, Lopez-Martinez MC (1998) Serum zinc and copper concentrations and Cu/Zn ratios in patients with hepatopathies or diabetes. J Trace Elem Med Biol 12:44–49. https://doi.org/10.1016/s0946-672x(98)80020-5
Hantke K (2001) Bacterial zinc transporters and regulators. Biometals 14:239–249. https://doi.org/10.1023/a:1012984713391
Pinilla-Tenas JJ, Sparkman BK, Shawki A, Illing AC, Mitchell CJ, Zhao N, Liuzzi JP, Cousins RJ, Knutson MD, Mackenzie B (2011) Zip14 is a complex broad-scope metal-ion transporter whose functional properties support roles in the cellular uptake of zinc and nontransferrin-bound iron. Am J Physiol Cell Physiol 301:C862-871. https://doi.org/10.1152/ajpcell.00479.2010
Wang CY, Jenkitkasemwong S, Duarte S, Sparkman BK, Shawki A, Mackenzie B, Knutson MD (2012) ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J Biol Chem 287:34032–34043. https://doi.org/10.1074/jbc.M112.367284
Lichten LA, Cousins RJ (2009) Mammalian zinc transporters: nutritional and physiologic regulation. Annu Rev Nutr 29:153–176. https://doi.org/10.1146/annurev-nutr-033009-083312
Zhao H, Eide D (1996) The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci U S A 93:2454–2458. https://doi.org/10.1073/pnas.93.6.2454
Grotz N, Fox T, Connolly E, Park W, Guerinot ML, Eide D (1998) Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc Natl Acad Sci U S A 95:7220–7224. https://doi.org/10.1073/pnas.95.12.7220
Taylor KM, Morgan HE, Johnson A, Hadley LJ, Nicholson RI (2003) Structure-function analysis of LIV-1, the breast cancer-associated protein that belongs to a new subfamily of zinc transporters. Biochem J 375:51–59. https://doi.org/10.1042/Bj20030478
Lin W, Chai J, Love J, Fu D (2010) Selective electrodiffusion of zinc ions in a Zrt-, Irt-like protein, ZIPB. J Biol Chem 285:39013–39020. https://doi.org/10.1074/jbc.M110.180620
Bin BH, Fukada T, Hosaka T, Yamasaki S, Ohashi W, Hojyo S, Miyai T, Nishida K, Yokoyama S, Hirano T (2011) Biochemical characterization of human ZIP13 protein: a homo-dimerized zinc transporter involved in the spondylocheiro dysplastic Ehlers-Danlos syndrome. J Biol Chem 286:40255–40265. https://doi.org/10.1074/jbc.M111.256784
Gupta S, Merriman C, Petzold CJ, Ralston CY, Fu D (2019) Water molecules mediate zinc mobility in the bacterial zinc diffusion channel ZIPB. J Biol Chem 294:13327–13335. https://doi.org/10.1074/jbc.RA119.009239
Zhang T, Liu J, Fellner M, Zhang C, Sui D, Hu J (2017) Crystal structures of a ZIP zinc transporter reveal a binuclear metal center in the transport pathway. Sci Adv 3:e1700344. https://doi.org/10.1126/sciadv.1700344
Eide DJ (2004) The SLC39 family of metal ion transporters. Pflug Arch 447:796–800. https://doi.org/10.1007/s00424-003-1074-3
Zhou JZ, He Q, Hemme CL, Mukhopadhyay A, Hillesland K, Zhou AF, He ZL, Van Nostrand JD, Hazen TC, Stahl DA, Wall JD, Arkin AP (2011) How sulphate-reducing microorganisms cope with stress: lessons from systems biology. Nat Rev Microbiol 9:452–466. https://doi.org/10.1038/nrmicro2575
Biswas KC, Woodards NA, Xu H, Barton LL (2009) Reduction of molybdate by sulfate-reducing bacteria. Biometals 22:131–139. https://doi.org/10.1007/s10534-008-9198-8
Cabrera G, Perez R, Gomez JM, Abalos A, Cantero D (2006) Toxic effects of dissolved heavy metals on Desulfovibrio vulgaris and Desulfovibrio sp. strains. J Hazard Mater 135:40–46. https://doi.org/10.1016/j.jhazmat.2005.11.058
Chardin B, Dolla A, Chaspoul F, Fardeau ML, Gallice P, Bruschi M (2002) Bioremediation of chromate: thermodynamic analysis of the effects of Cr(VI) on sulfate-reducing bacteria. Appl Microbiol Biotechnol 60:352–360. https://doi.org/10.1007/s00253-002-1091-8
Goulhen F, Gloter A, Guyot F, Bruschi M (2006) Cr(VI) detoxification by Desulfovibrio vulgaris strain Hildenborough: microbe-metal interactions studies. Appl Microbiol Biotechnol 71:892–897. https://doi.org/10.1007/s00253-005-0211-7
Humphries AC, Nott KP, Hall LD, Macaskie LE (2004) Continuous removal of Cr(VI) from aqueous solution catalysed by palladised biomass of Desulfovibrio vulgaris. Biotechnol Lett 26:1529–1532. https://doi.org/10.1023/B:BILE.0000044457.80314.4d
Klonowska A, Clark ME, Thieman SB, Giles BJ, Wall JD, Fields MW (2008) Hexavalent chromium reduction in Desulfovibrio vulgaris Hildenborough causes transitory inhibition of sulfate reduction and cell growth. Appl Microbiol Biotechnol 78:1007–1016. https://doi.org/10.1007/s00253-008-1381-x
Ma C, Hao Z, Huysmans G, Lesiuk A, Bullough P, Wang Y, Bartlam M, Phillips SE, Young JD, Goldman A, Baldwin SA, Postis VL (2015) A versatile strategy for production of membrane proteins with diverse topologies: application to investigation of bacterial homologues of human divalent metal ion and nucleoside transporters. PLoS ONE 10:e0143010. https://doi.org/10.1371/journal.pone.0143010
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Davis IW, Murray LW, Richardson JS, Richardson DC (2004) MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32:W615-619. https://doi.org/10.1093/nar/gkh398
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375-383. https://doi.org/10.1093/nar/gkm216
Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. https://doi.org/10.1107/S0907444909042073
Janson G, Zhang C, Prado MG, Paiardini A (2017) PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. Bioinformatics 33:444–446. https://doi.org/10.1093/bioinformatics/btw638
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30(Suppl 1):S162-173. https://doi.org/10.1002/elps.200900140
Whelan S, Goldman N (2001) A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. https://doi.org/10.1093/oxfordjournals.molbev.a003851
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. https://doi.org/10.1093/bioinformatics/8.3.275
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Postis VLG, Deacon SE, Roach PCJ, Wright GSA, Xia X, Ingram JC, Hadden JM, Henderson PJF, Phillips SEV, McPherson MJ, Baldwin SA (2008) A high-throughput assay of membrane protein stability. Mol Membr Biol 25:617–624. https://doi.org/10.1080/09687680802530469
Chao Y, Fu D (2004) Kinetic study of the antiport mechanism of an Escherichia coli zinc transporter, ZitB. J Biol Chem 279:12043–12050. https://doi.org/10.1074/jbc.M313510200
Guerinot ML (2000) The ZIP family of metal transporters. Biochim Biophys Acta 1465:190–198. https://doi.org/10.1016/s0005-2736(00)00138-3
Bernsel A, Viklund H, Hennerdal A, Elofsson A (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res 37:W465-468. https://doi.org/10.1093/nar/gkp363
Viklund H, Elofsson A (2004) Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci 13:1908–1917. https://doi.org/10.1110/ps.04625404
Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315
Jones DT (2007) Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23:538–544. https://doi.org/10.1093/bioinformatics/btl677
Gong CX, Ma C (2020) Detergents properties and their applications in membrane protein structural biology. Chin J Biochem Mol Biol 36:14–20
Lu M, Chai J, Fu D (2009) Structural basis for autoregulation of the zinc transporter YiiP. Nat Struct Mol Biol 16:1063-U1081. https://doi.org/10.1038/nsmb.1662
Wei YN, Fu D (2006) Binding and transport of metal ions at the dimer interface of the Escherichia coli metal transporter YiiP. J Biol Chem 281:23492–23502. https://doi.org/10.1074/jbc.M602254200
Lopez-Redondo ML, Coudray N, Zhang ZN, Alexopoulos J, Stokes DL (2018) Structural basis for the alternating access mechanism of the cation diffusion facilitator YiiP. Proc Natl Acad Sci USA 115:3042–3047. https://doi.org/10.1073/pnas.1715051115
Acknowledgements
We thank Dr Gerard Huysmans and other colleges for their kind supervision during this study and other related studies. As a friend of Prof. Steve Baldwin, I dedicate this paper to him in memoriam.
Funding
This work was originally supported by funds from Prof. Steve Baldwin, University of Leeds, UK. To complete this work, it was latter supported by grants from the Zhejiang Province Science and Technology Plan of Traditional Chinese Medicine [grant number 2020ZQ031], the Natural Science Foundation of China [grant number 32000707], and Zhejiang University [grant number SJS202014].
Author information
Authors and Affiliations
Contributions
CM designed the project, did the experiments, and performed the analysis. CM drafted the manuscript. CXG optimized the manuscript and provided partial funding support.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ma, C., Gong, C. Expression, Purification and Characterization of a ZIP Family Transporter from Desulfovibrio vulgaris. Protein J 40, 776–785 (2021). https://doi.org/10.1007/s10930-021-10008-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-021-10008-7