Abstract
Hyper-IgE syndromes (HIES) are a group of inborn errors of immunity (IEI) caused by monogenic defects such as in the gene STAT3 (STAT3-HIES). Patients suffering from HIES show an increased susceptibility to Staphylococcus aureus (S. aureus) including skin abscesses and pulmonary infections. To assess if the underlying immune defect of STAT3-HIES patients influences the resistance patterns, pathogenicity factors or strain types of S. aureus. We characterized eleven S. aureus strains isolated from STAT3-HIES patients (n = 4) by whole genome sequencing (WGS) to determine presence of resistance and virulence genes. Additionally, we used multi-locus sequence typing (MLST) and protein A (spa) typing to classify these isolates. Bacterial isolates collected from this cohort of STAT3-HIES patients were identified as common spa types in Germany. Only one of the isolates was classified as methicillin-resistant S. aureus (MRSA). For one STAT3 patient WGS illustrated that infection and colonization occurred with different S. aureus isolates rather than one particular clone. The identified S. aureus carriage profile on a molecular level suggests that S. aureus strain type in STAT3-HIES patients is determined by local epidemiology rather than the underlying immune defect highlighting the importance of microbiological assessment prior to antibiotic treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
While Staphylococcus aureus (S. aureus) frequently colonizes skin and mucosa, it is also a major pathogen causing harmful infections such as abscesses, pneumonia, and septicemia [1]. In fact, S. aureus is the leading cause for surgical site infections and especially methicillin-resistant S. aureus (MRSA) infections are associated with significant costs for health care systems [2].
Hyper-IgE syndromes (HIES) are a group of inborn errors of immunity (IEI), of which the most frequent form is caused by monogenic defects in the gene STAT3 (STAT3-HIES) [3,4,5]. STAT3-HIES is characterized by elevated serum IgE levels, eosinophilia, eczema and increased susceptibility to S. aureus infections [4,5,6]. Severe staphylococcal skin and lung infections are common in STAT3-HIES patients, most likely because of the impairment in the epithelial immune response [7,8,9]. S. aureus can also cause infections in immunocompetent individuals; the predisposition of STAT3-HIES patients towards this pathogen, however, offers further insights into the immune defense against S. aureus. STAT3-HIES patients show impaired Th17 cell function and diminished memory B cell development [10,11,12,13]. Additionally, STAT3-HIES patients fail to raise antibodies against S. aureus toxin despite chronic colonization [14]. Importantly, besides an impaired humoral and cellular immune response towards S. aureus, STAT3-HIES patients also suffer from reduced epithelial immunity towards this pathogen [15]. STAT3-HIES patients benefit from Immunoglobulin replacement therapy (IgRT), most likely because it partly compensates the compromised humoral and cellular immune response [16, 17]. Nevertheless, new therapeutic strategies are needed to improve the clinical outcome of patients.
Although S. aureus colonization and infection are key symptoms of STAT3-HIES and patients frequently require antibiotic prophylaxis or therapy, little is known about the molecular features of S. aureus in these patients. Moreover, it is unclear if the immunodeficiency of STAT3-HIES patients selects S. aureus strains with a specific “genetic fingerprint”. A recent study used multi-locus sequence typing (MLST) and protein A (spa) typing to analyze the genetic background of 13 S. aureus isolates collected from STAT3-HIES patients in the USA [8]. Here, most isolates resembled highly virulent USA300 strains that are prevalent in hospitals in the USA and have been associated with MRSA outbreaks. The aim of our study was to analyze the S. aureus carriage profile of STAT3-HIES patients in Germany to optimize antibiotic treatment using bacterial whole genome sequencing (WGS) and to investigate virulence and resistance genes in these isolates.
Methods
Patients
All four STAT3-HIES patients carried a heterozygous dominant-negative STAT3 mutation, were unrelated, and presented with the characteristic clinical findings of their genetically confirmed diagnosis at inclusion of study (Table 1). Mutations were reported using the nomenclature of den Dunnen and Antonarakis [19].
S. aureus Isolate Collection, Whole Genome Sequencing, and Data Analysis
S. aureus strains were collected and cultured using standard procedures. In total eleven S. aureus isolates were collected including screening isolates (nose/throat n = 4; perianal n = 1) isolates collected from skin lesions (n = 5) and one clinical isolate from a lymph node abscess (please see Table 2 for further isolate and collection site details). Genomic DNA of S. aureus isolates was purified using the MagAttract HMW DNA kit (Qiagen, Venlo Netherlands) following manufacturer’s instructions. Isolates were sequenced using Illumina technology and Nextera XT version 2 chemistry, with a 250-bp paired-end protocol on a MiSeq sequencer (Illumina, San Diego, USA). Quality trimming of fastq files (average base quality of 30, aiming for 100-fold coverage) and de novo assembly using SKESA [20] were performed with SeqSphere + (version 6; Ridom GmbH, Münster, Germany) [21]. Only genomes harboring ≥ 95% core genome multi-locus sequence typing (cgMLST) targets of the S. aureus cgMLST scheme passed quality control; otherwise, sequencing was repeated. Target gene sets for virulence factors, resistance, and toxin genes as well as spa type were analyzed using the SeqSphere + software [21, 22]. Presence of enterotoxin genes was confirmed by polymerase chain reaction as described previously [23].
Results
Patient Characteristics and S. aureus Isolates
Patients included in our study were between 5 and 53 years of age at enrollment. All four patients had a history of pulmonary and skin infections (Table 1). During the study period, patients received prophylactic antibiotic treatment as specified in Table 1 and skin treatment with octenidine dihydrochloride as well as symptomatic skin care with emollients. Eleven S. aureus isolates were collected either as screening isolates (nasal carriage) or from different skin sites. One clinical isolate (P1.2) was isolated from a lymph node abscess.
Epidemiology of S. aureus Strains and spa Typing Results
All bacteria were analyzed for MLST, clonal complex (CC) and spa type. The collected S. aureus isolates were predominantly ST5 or ST582 and the most frequent spa types were t179 and t084 (Table 2). Only one isolate out of eleven was identified as MRSA. In addition, none of the isolates carried the Panton-Valentine leukocidin (pvl), staphylococcal enterotoxins Q (seq), or staphylococcal enterotoxins K (sek) gene (Tables 2 and 3). These typing results match the general epidemiology of S. aureus in Germany and differ from previous reports that identified mostly MRSA isolates encoding the pvl gene in STAT3-HIES patients from the USA [24].
Clonal Relationship of S. aureus Isolates in STAT3-HIES Patients
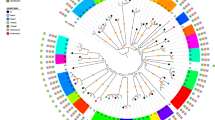
Next, the genetic relatedness of S. aureus isolates collected from STAT3-HIES patients was analyzed by cgMLST. cgMLST combines “classical” typing approaches to classify bacteria with the extensive genetic data sets obtained by WGS [25]. For S. aureus up to 1861 conserved target genes are compared to discriminate between isolates achieving a sufficient resolution for epidemiological and surveillance studies [26]. The isolates collected as part of this study were genetically unrelated and differed significantly from a reference sequence of highly virulent USA300 (NC_007793.1) (Fig. 1a). In one STAT3-HIES patient (P3) five S. aureus isolates were isolated from skin lesions at different sites yet at the same time point (P3.1 − P3.5). WGS showed that these isolates belonged to the same ST (ST582) and spa type (t084) but differed by up to 6 alleles or 6 single nucleotide polymorphisms (SNPs) in their core genome (Fig. 1b). In another STAT3-HIES patient (P1) WGS illustrated that the S. aureus isolate causing a lymph node infection (P1.2) was not genetically related to the clones colonizing the patient at other sites and differed from colonizing isolates in 1401 alleles or 9382 SNPs (Fig. 1c). The colonizing isolates P1.1, P1.3, and P1.4 belong to the same genetic family, ST (ST5) and spa type (t179), yet showed significant variation in their core genome. Strains P1.1 and P1.3 were both isolated from a nose/throat screening swab but were collected 8 years apart and differed by 30 alleles or 31 SNPs in their core genome. Taken together the analyses of these bacterial isolates indicate that STAT3-HIES patients are carrying several genetically diverse S. aureus isolates at a time.
Minimum spanning trees of S. aureus isolates illustrate their genotypic relationship. Minimum spanning trees were based on up to 1861 cgMLST target genes, pairwise ignoring missing values. Every circle represents one genotype while connecting lines represent the number of different alleles in a pairwise comparison. a S. aureus isolates of different STAT3-HIES patients (P1–P4, using a different color for each patient). A reference sequence of a reference USA300 strain (NC_007793.1) was included as comparison (white). b The skin colonizing isolates of STAT3-HIES patient P3 (blue). c The isolate causing a lymph node abscess in STAT3-HIES patient P1 (P1.2, dark red) in comparison to three other isolates of the same patient. Isolates P1.1 and P1.3 are both isolated from a nose/throat screening swab (light red) but were collected 8 years apart. Isolate P1.4 (pink, perianal screening swab) has been collect at the same time point as P1.3
Pathogenicity Factors in Patients’ S. aureus Strains
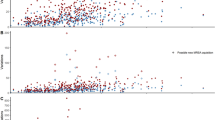
Several studies have correlated the ability of S. aureus to cause soft tissue infection and inflammation with the activity of pathogenicity factors and bacterial toxin genes [22]. Therefore, we analyzed the S. aureus isolates of STAT3-HIES patients for genes of the accessory regulator (agr), hemolysin genes (hl), and genes encoding for enterotoxins, adhesion molecules and the immune evasion complex (IEC) (Table 3). In addition, we constructed a unweighted pair group method with arithmetic mean (UPGMA)-tree based on their toxin and virulence gene profile of the S. aureus isolates to illustrate the distribution pattern of the 45 pathogenicity factors analyzed in the study (Fig. 2).
UPGMA-tree based on the toxin and virulence gene profile of eleven S. aureus isolates. The tree was drawn to scale with branches given in absolute alleles distance, using a different color for each patient (colors correspond to Fig. 1)
Hemolysin α (hla) was present in all isolates. Two colonizing isolates showed a stop codon in the hl gene (P1.3 and P1.4). One colonizing isolate lacked the hemolysin δ (hld) gene (P3.4). The IEC consists of the genes coding for staphylokinase (sak), staphylococcal complement inhibitor (scn), chemotaxis inhibitory protein (chp), and staphylococcal enterotoxins A (sea). IEC genes are well-known pathogenicity factors that aid S. aureus to bypass the human immune response by counteracting key steps in innate immunity such as complement activation and chemotaxis. Interestingly, three of the eleven isolates collected from STAT3-HIES patients contained all four IEC genes (isolates P1.1, P1.3 and P1.4, all isolated from patient P1). As STAT3-HIES patients show reduced Th17 and B cell responses towards S. aureus, bacterial pathogenicity factors that impair innate immunity might result in a more pronounced effect in these patients. However, long-term follow-up studies are needed to address this research question.
Discussion
Presently, the molecular characteristics and genetic variability of S. aureus clones in STAT3-HIES patients remain largely unknown. The MLST ST and spa types of the eleven S. aureus isolates collected during this study differed significantly from the typing results described in a previous report from the USA [18]. However, the identified ST and spa types are common in Germany [22). The ST and spa type results imply that the local epidemiology is the main factor determining which S. aureus strains colonize STAT3-HIES patients. Although all patients enrolled in our study have received long-time antibiotic prophylaxis and therapy, MRSA was not prevalent in our patient cohort (Table 2). In fact, MRSA infections in the healthcare setting have been decreasing steadily over the recent years in Germany [27, 28]. In 2018, a surveillance conducted by Germany’s public health institution, the Robert Koch-Institute, estimated that 7.7% of all S. aureus bacteria isolated from patients in the community setting are MRSA [29]. Further studies are required to investigate if the prevalence of MRSA among STAT3-HIES patients equals the prevalence of the general population in their home countries. Yet at present, we recommend that empirical antibiotic treatment for severe infection in STAT3-HIES patients might not have to cover MRSA unless a patient is a known carrier or the local MRSA prevalence is high. Despite the limitation of relative low patient number of patients enrolled, due to STAT3-HIES being a rare disease, our study shows first interesting results how WGS can be used to characterize S. aureus isolates collected from STAT3-HIES patients. In our study, we analyzed the genetic relatedness of colonizing S. aureus isolates from several STAT3-HIES patients based on WGS data. Here, we demonstrated that STAT3-HIES patients were colonized with genetically diverse S. aureus clones at a time. Similar observations have been made in a study analyzing S. aureus isolates collected from eczematic lesions of atopic eczema patients by WGS. In this publication, a broad genetic diversity of bacterial isolates was detected suggesting that clonal expansion of a bacterial population takes place during a disease flare [30].
The WGS data of S. aureus isolates generated as part of this study offer a comprehensive characterization of multiple virulence factors. An improved understanding of pathogenicity factors may pave the way for new therapeutic strategies for STAT3-HIES patients. Currently, long-acting monoclonal antibodies that are capable to neutralize S. aureus toxins in the respiratory tract are under development to treat MRSA lung infections [31, 32]. As STAT3-HIES patients show reduced IgG levels for S. aureus toxins, the potential of these monoclonal antibodies as treatment for severe S. aureus lung infections could be evaluated [14, 33].
Taken together, our study provides a detailed view into the molecular characteristics of the S. aureus isolates collected from STAT3-HIES patients, including genetic relatedness of isolates, methicillin resistance and presence of virulence factors and demonstrates the benefits of using molecular approaches to study host–pathogen dynamics in IEI.
Data Availability
The whole genome datasets of S. aureus isolates generated for this study have been deposited in the European Nucleotide Archive (ENA) at EMBL-EBI under accession number PRJEB52061 (https://www.ebi.ac.uk/ena/browser/view/PRJEB52061).
Code Availability
The code related to data analysis is available from the corresponding author on request.
Abbreviations
- agr :
-
Accessory gen regulator
- cgMLST:
-
Core genome multi-locus sequence typing
- chp :
-
Chemotaxis inhibitory protein of staphylococcus
- CC:
-
Clonal complex
- HIES:
-
Hyper-IgE syndromes
- hl :
-
Hemolysin
- hla :
-
Hemolysin α
- hld :
-
Hemolysin δ
- IEC:
-
Immune evasion cluster
- IgRT:
-
Immunoglobulin replacement therapy
- IEI:
-
Inborn error of immunity
- MRSA:
-
Methicillin-resistant S. aureus
- MLST:
-
Multi-locus sequence typing
- pvl :
-
Panton-Valentine leukocidin
- spa :
-
Protein A
- scn :
-
Staphylococcal complement inhibitor
- sea :
-
Staphylococcal enterotoxins A
- sek :
-
Staphylococcal enterotoxins K
- seq :
-
Staphylococcal enterotoxins Q
- S. aureus :
-
Staphylococcus aureus
- sak :
-
Staphylokinase
- UPGMA:
-
Unweighted pair group method with arithmetic mean
- WGS:
-
Whole genome sequencing
References
Krismer B, Weidenmaier C, Zipperer A, Peschel A. The commensal lifestyle of Staphylococcus aureus and its interactions with the nasal microbiota. Nat Rev Microbiol. 2017;15(11):675–87.
Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370(13):1198–208.
Holland SM, DeLeo FR, Elloumi HZ, Hsu AP, Uzel G, Brodsky N, et al. STAT3 mutations in the hyper-IgE syndrome. N Engl J Med. 2007;357(16):1608–19.
Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, et al. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448(7157):1058–62.
Renner ED, Torgerson TR, Rylaarsdam S, Añover-Sombke S, Golob K, LaFlam T, et al. STAT3 mutation in the original patient with Job’s syndrome. N Engl J Med. 2007;357(16):1667–8.
Freeman AF, Kleiner DE, Nadiminti H, Davis J, Quezado M, Anderson V, et al. Causes of death in hyper-IgE syndrome. J Allergy Clin Immunol. 2007;119(5):1234–40.
Minegishi Y, Saito M, Nagasawa M, Takada H, Hara T, Tsuchiya S, et al. Molecular explanation for the contradiction between systemic Th17 defect and localized bacterial infection in hyper-IgE syndrome. J Exp Med. 2009;206(6):1291–301.
Kröner C, Neumann J, Ley-Zaporozhan J, Hagl B, Meixner I, Spielberger BD, et al. Lung disease in STAT3 hyper-IgE syndrome requires intense therapy. Allergy. 2019;74(9):1691–702.
Hagl B, Heinz V, Schlesinger A, Spielberger BD, Sawalle-Belohradsky J, Senn-Rauh M, et al. Key findings to expedite the diagnosis of hyper-IgE syndromes in infants and young children. Pediatr Allergy Immunol. 2016;27(2):177–84.
Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, et al. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452(7188):773–6.
van de Veen W, Krätz CE, McKenzie CI, Aui PM, Neumann J, van Noesel CJM, et al. Impaired memory B-cell development and antibody maturation with a skewing toward IgE in patients with STAT3 hyper-IgE syndrome. Allergy. 2019;74(12):2394–405.
Renner ED, Rylaarsdam S, Anover-Sombke S, Rack AL, Reichenbach J, Carey JC, et al. Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J Allergy Clin Immunol. 2008;122(1):181–7.
Meyer-Bahlburg A, Renner ED, Rylaarsdam S, Reichenbach J, Schimke LF, Marks A, et al. Heterozygous signal transducer and activator of transcription 3 mutations in hyper-IgE syndrome result in altered B-cell maturation. J Allergy Clin Immunol. 2012;129(2):559–62, 562.e1-2.
Stentzel S, Hagl B, Abel F, Kahl BC, Rack-Hoch A, Bröker BM, et al. Reduced Immunoglobulin (Ig) G Response to Staphylococcus aureus in STAT3 Hyper-IgE Syndrome. Clin Infect Dis. 2017;64(9):1279–82.
Myles IA, Anderson ED, Earland NJ, Zarember KA, Sastalla I, Williams KW, et al. TNF overproduction impairs epithelial staphylococcal response in hyper IgE syndrome. J Clin Invest. 2018;128(8):3595–604.
Kimata H. High-dose intravenous gamma-globulin treatment for hyperimmunoglobulinemia E syndrome. J Allergy Clin Immunol. 1995;95(3):771–4.
Chandesris M-O, Melki I, Natividad A, Puel A, Fieschi C, Yun L, et al. Autosomal dominant STAT3 deficiency and hyper-IgE syndrome: molecular, cellular, and clinical features from a French national survey. Medicine (Baltimore). 2012;91(4):e1-19.
Sastalla I, Williams KW, Anderson ED, Myles IA, Reckhow JD, Espinoza-Moraga M, et al. Molecular typing of staphylococcus aureus isolated from patients with autosomal dominant hyper IgE syndrome. Pathogens. 2017;6(2):23. https://doi.org/10.3390/pathogens6020023.
den Dunnen JT, Antonarakis SE. Nomenclature for the description of human sequence variations. Hum Genet. 2001;109(1):121–4.
Souvorov A, Agarwala R, Lipman DJ. SKESA: strategic k-mer extension for scrupulous assemblies. Genome Biol. 2018;19(1):153.
Leopold SR, Goering RV, Witten A, Harmsen D, Mellmann A. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol. 2014;52(7):2365–70.
Lebughe M, Phaku P, Niemann S, Mumba D, Peters G, Muyembe-Tamfum J-J, et al. The impact of the staphylococcus aureus virulome on infection in a developing country: a cohort study. Front Microbiol. 2017;8:1662.
Omoe K, Hu D-L, Takahashi-Omoe H, Nakane A, Shinagawa K. Comprehensive analysis of classical and newly described staphylococcal superantigenic toxin genes in Staphylococcus aureus isolates. FEMS Microbiol Lett. 2005;246(2):191–8.
Becker K, Schaumburg F, Fegeler C, Friedrich AW, Köck R. Staphylococcus aureus from the German general population is highly diverse. Int J Med Microbiol. 2017;307(1):21–7.
Schürch AC, Arredondo-Alonso S, Willems RJL, Goering RV. Whole genome sequencing options for bacterial strain typing and epidemiologic analysis based on single nucleotide polymorphism versus gene-by-gene-based approaches. Clin Microbiol Infect. 2018;24(4):350–4.
Mellmann A, Bletz S, Böking T, Kipp F, Becker K, Schultes A, et al. Real-time genome sequencing of resistant bacteria provides precision infection control in an institutional setting. J Clin Microbiol. 2016;54(12):2874–81.
Meyer E, Schröder C, Gastmeier P, Geffers C. The reduction of nosocomial MRSA infection in Germany: an analysis of data from the Hospital Infection Surveillance System (KISS) between 2007 and 2012. Dtsch Arztebl Int. 2014;111(19):331–6.
Kramer TS, Schröder C, Behnke M, Aghdassi SJ, Geffers C, Gastmeier P, et al. Decrease of methicillin resistance in Staphylococcus aureus in nosocomial infections in Germany-a prospective analysis over 10 years. J Infect. 2019;78(3):215–9.
Layer F, Strommenger B, Cuny C, Noll I, Eckmanns T, Werner G. Eigenschaften, Häufigkeit und Verbreitung von MRSA in Deutschland. Epid Bull. 2019;42:437–42. https://doi.org/10.25646/6320.2.
Harkins CP, Pettigrew KA, Oravcová K, Gardner J, Hearn RMR, Rice D, et al. The microevolution and epidemiology of staphylococcus aureus colonization during atopic eczema disease flare. J Invest Dermatol. 2018;138(2):336–43.
Diep BA, Le VTM, Badiou C, Le HN, Pinheiro MG, Duong AH, et al. IVIG-mediated protection against necrotizing pneumonia caused by MRSA. Sci Transl Med. 2016;8(357):357ra124.
Ruzin A, Wu Y, Yu L, Yu X-Q, Tabor DE, Mok H, et al. Characterisation of anti-alpha toxin antibody levels and colonisation status after administration of an investigational human monoclonal antibody, MEDI4893, against Staphylococcus aureus alpha toxin. Clin Trans Immunol. 2018;7(1): e1009.
Hua L, Hilliard JJ, Shi Y, Tkaczyk C, Cheng LI, Yu X, et al. Assessment of an anti-alpha-toxin monoclonal antibody for prevention and treatment of Staphylococcus aureus-induced pneumonia. Antimicrob Agents Chemother. 2014;58(2):1108–17.
Acknowledgements
We thank all patients and their referring physicians for their participation. We thank the sequencing team of the Institute of Hygiene at University Münster, the team of the Microbiology Laboratory of Dr. von Haunersches Kinderspital LMU, as well as Daniela Kreilinger, Translational Immunology in Environmental Medicine, School of Medicine at TUM and Amadeo de Tomassi, Chair of Environmental Medicine, University of Augsburg for excellent technical assistance. In addition, we thank Prof. Barbara Kahl, Institute of Medical Microbiology, University Hospital Münster and Prof. Stefanie Kampmeier, Institute of Hygiene, University Hospital Münster for critical review of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the German Research Foundation DFG RE2799/6–1 (EDR), the Job Research Foundation (VS and EDR) and LMU Munich FöFoLe (FA and EDR), the Helmholtz-Future topic “Immunology and Inflammation” (ZT-0027).
Author information
Authors and Affiliations
Contributions
Conceptualization and methodology: VS, EDR, and AM. Validation: VS. Formal analysis: VS. Funding acquisition: VS and EDR. Investigation: VS, RE, FA, GN, SD, BH, and EDR. Resources and patient material: VS, FA, GN, JH, VSI, RH, JHU, AM, and EDR. Data Curation: RE, MR, and BH. Writing—original draft preparation: VS. Writing—review and editing: VS, RE, MR, BH, AM, and EDR. All authors have accepted responsibility for the entire content of this manuscript and approved its submission and publication.
Corresponding author
Ethics declarations
Ethics Approval
The study was approved by the Institutional Review Board of the Medical School TUM (32/20S-KH).
Consent to Participate
Written informed consent was obtained from all patients or their legal guardians.
Consent for Publication
Patients signed informed consent regarding publishing their data.
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Schwierzeck, V., Effner, R., Abel, F. et al. Molecular Assessment of Staphylococcus Aureus Strains in STAT3 Hyper-IgE Syndrome Patients. J Clin Immunol 42, 1301–1309 (2022). https://doi.org/10.1007/s10875-022-01293-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-022-01293-7