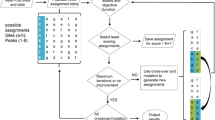
Abstract
A strategy for acquiring structural information from sparsely isotopically labeled large proteins is illustrated with an application to the E. coli heat-shock protein, HtpG (high temperature protein G), a 145 kDa dimer. It uses 13C-alanine methyl labeling in a perdeuterated background to take advantage of the sensitivity and resolution of Methyl-TROSY spectra, as well as the backbone-centered structural information from 1H–13C residual dipolar couplings (RDCs) of alanine methyl groups. In all, 40 of the 47 expected crosspeaks were resolved and 36 gave RDC data. Assignments of crosspeaks were partially achieved by transferring assignments from those made on individual domains using triple resonance methods. However, these were incomplete and in many cases the transfer was ambiguous. A genetic algorithm search for consistency between predictions based on domain structures and measurements for chemical shifts and RDCs allowed 60% of the 40 resolved crosspeaks to be assigned with confidence. Chemical shift changes of these crosspeaks on adding an ATP analog to the apo-protein are shown to be consistent with structural changes expected on comparing previous crystal structures for apo- and complex- structures. RDCs collected on the assigned alanine methyl peaks are used to generate a new solution model for the apo-protein structure.







Similar content being viewed by others
References
Acton TB et al (2011) Preparation of protein samples for NMR structure, function, and small-molecule screening studies. In: Kuo LC (ed) Fragment-based drug design: tools, practical approaches, and examples, vol 493. Methods in enzymology. Academic Press, Amsterdam, pp 21–60. doi:10.1016/b978-0-12-381274-2.00002-9
Ali MMU et al (2006) Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440:1013–1017. doi:10.1038/nature04716
Allegrozzi M, Bertini I, Janik MBL, Lee YM, Lin GH, Luchinat C (2000) Lanthanide induced pseudocontact shifts for solution structure refinements of macromolecules in shells up to 40 Å from the metal ion. J Am Chem Soc 122:4154–4161
Ayala I, Sounier R, Use N, Gans P, Boisbouvier J (2009) An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J Biomol NMR 43:111–119. doi:10.1007/s10858-008-9294-7
Ayala I, Hamelin O, Amero C, Pessey O, Plevin MJ, Gans P, Boisbouvier J (2012) An optimized isotopic labelling strategy of isoleucine-gamma(2) methyl groups for solution NMR studies of high molecular weight proteins. Chem Commun 48:1434–1436. doi:10.1039/c1cc12932e
Bahrami A, Assadi A, Markley JL, Eghbalnia H (2009) Probablilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectrscopy. PLoS Comput Biol 5:e1000307
Battiste JL, Wagner G (2000) Utilization of site-directed spin labeling and high resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. BioChemistry 39:5355–5365
Bax A (2003) Weak alignment offers new NMR opportunities to study protein structure and dynamics. Protein Sci 12:1–16. doi:10.1110/ps.0233303
Burmann BM, Hiller S (2015) Chaperones and chaperone-substrate complexes: dynamic playgrounds for NMR spectroscopists. Prog Nucl Magn Reson Spectrosc 86–87:41–64. doi:10.1016/j.pnmrs.2015.02.004
Clore GM (2011) Exploring sparsely populated states of macromolecules by diamagnetic and paramagnetic NMR relaxation. Protein Sci 20:229–246
Day JEH et al (2011) Targeting the Hsp90 molecular chaperone with novel macrolactams. Synthesis, structural, binding, and cellular studies. Acs Chem Biol 6:1339–1347 doi:10.1021/cb200196e
Didenko T, Duarte AMS, Karagoz GE, Rudiger SGD (2012) Hsp90 structure and function studied by NMR spectroscopy. Biochim Biophys Acta 1823:636–647. doi:10.1016/j.bbamcr.2011.11.009
Dollins DE, Warren JJ, Immormino RM, Gewirth DT (2007) Structures of Grp94-nucleotide complexes reveal mechanistic differences between the Hsp90 chaperones. Mol Cell 28:41–56. doi:10.1016/j.molcel.2007.08.024
Gao Q et al (2016) Structural aspects of heparan sulfate binding to Robo1-Ig1-2. ACS Chem Biol 11:3106–3113
Gao Q, Chalmers GR, Moremen KM, Prestegard JH (2017) NMR assignments of sparsely labeled proteins using a genetic algorithm. J Biomol NMR 67:283–294. doi:10.1007/s10858-017-0101-1
Godoy-Ruiz R, Guo CY, Tugarinov V (2010) Alanine methyl groups as NMR probes of molecular structure and dynamics in high-molecular-weight proteins. J Am Chem Soc 132:18340–18350. doi:10.1021/ja1083656
Harris SF, Shiau AK (2004) The crystal structure of the carboxy-terminal dimerization domain of HtpG, the Escherichia coli Hsp90, reveals a potential substrate binding site. Structure 12:1087–1097
Hennig J, Sattler M (2014) The dynamic duo: combining NMR and small angle scattering in structural biology. Protein Sci 23:669–682. doi:10.1002/pro.2467
Hogben HJ, Krzystyniak M, Charnock GTP, Hore PJ, Kuprov I (2011) Spinach—a software library for simulation of spin dynamics in large spin systems. J Magn Reson 208:179–194. doi:10.1016/j.jmr.2010.11.008
Isaacson RL, Simpson PJ, Liu M, Cota E, Zhang X, Freemont P, Matthews S (2007) A new labeling method for methyl transverse relaxation-optimized spectroscopy NMR spectra of alanine residues. J Am Chem Soc 129:15428-+ doi:10.1021/ja0761784
Jackson SE (2013) Hsp90: structure and function. In: Jackson S (ed) Molecular chaperones, vol 328. Topics in current chemistry. Springer, Berlin, pp 155–240. doi:10.1007/128_2012_356
Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B (1996) High-level production of uniformly N-15- and C-13-enriched fusion proteins in Escherichia coli. J Biomol NMR 7:131–141
Karagoz GE et al (2011) N-terminal domain of human Hsp90 triggers binding to the cochaperone p23. Proc Natl Acad Sci USA 108:580–585. doi:10.1073/pnas.1011867108
Karagoz GE et al (2014) Hsp90-tau complex reveals molecular basis for specificity in chaperone action. Cell 156:963–974. doi:10.1016/j.cell.2014.01.037
Kay LE, Gardner KH (1997) Solution NMR spectroscopy beyond 25 kda. Curr Opin Struct Biol 7:722–731. doi:10.1016/s0959-440x(97)80084-x
Krukenberg KA, Forster F, Rice LM, Sali A, Agard DA (2008) Multiple conformations of E. coli Hsp90 in solution: insights into the conformational dynamics of Hsp90. Structure 16:755–765. doi:10.1016/j.str.2008.01.021
Krukenberg KA, Bottcher UMK, Southworth DR, Agard DA (2009) Grp94, the endoplasmic reticulum Hsp90, has a similar solution conformation to cytosolic Hsp90 in the absence of nucleotide. Protein Sci 18:1815–1827. doi:10.1002/pro.191
Krukenberg KA, Street TO, Lavery LA, Agard DA (2011) Conformational dynamics of the molecular chaperone Hsp90. Q Rev Biophys 44:229–255. doi:10.1017/s0033583510000314
Lavery LA, Partridge JR, Ramelot TA, Elnatan D, Kennedy MA, Agard DA (2014) Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism. Mol Cell 53:330–343. doi:10.1016/j.molcel.2013.12.023
Li D, Brüschweiler R (2015) PPM_One: a static protein structure based chemical shift predictor. J Biomol NMR 62:403–409
Lipsitz RS, Tjandra N (2004) Residual dipolar couplings in NMR structure analysis. Annu Rev Biophys Biomol Struct 33:387–413
Miyata Y, Nakamoto H, Neckers L (2013) The therapeutic target Hsp90 and cancer hallmarks. Curr Pharm Des 19:347–365
Nietlispach D et al (1996) An approach to the structure determination of larger proteins using triple resonance NMR experiments in conjunction with random fractional deuteration. J Am Chem Soc 118:407–415. doi:10.1021/ja952207b
Ollerenshaw JE, Tugarinov V, Kay LE (2003) Methyl TROSY: explanation and experimental verification. Magn Reson Chem 41:843–852. doi:10.1002/mrc.1256
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi:10.1002/jcc.20084
Prestegard JH, Al-Hashimi HM, Tolman JR (2000) NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys 33:371–424. doi:10.1017/s0033583500003656
Prestegard JH, Bougault CM, Kishore AI (2004) Residual dipolar couplings in structure determination of biomolecules. Chem Rev 104:3519–3540. doi:10.1021/cr030419i
Retzlaff M et al (2010) Asymmetric activation of the Hsp90 dimer by its cochaperone aha1. Mol Cell 37:344–354. doi:10.1016/j.molcel.2010.01.006
Rosenzweig R, Kay LE (2014) Bringing dynamic molecular machines into focus by methyl-trosy NMR. Annu Rev Biochem 83:291–315. doi:10.1146/annurev-biochem-060713-035829
Rosenzweig R, Kay LE (2016) Solution NMR spectroscopy provides an avenue for the study of functionally dynamic molecular machines: the example of protein disaggregation. J Am Chem Soc 138:1466–1477. doi:10.1021/jacs.5b11346
Sali A, Blundell TL (1993) Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol 234:779–815. doi:10.1006/jmbi.1993.1626
Schmitz C, Vernon R, Otting G, Baker D, Huber T (2012) Protein structure determination from pseudocontact shifts using rosetta. J Mol Biol 416:668–677
Schulze A, Beliu G, Helmerich DA, Schubert J, Pearl LH, Prodromou C, Neuweiler H (2016) Cooperation of local motions in the Hsp90 molecular chaperone atpase mechanism. Nat Chem Biol 12:628–635. doi:10.1038/nchembio.2111
Shiau AK, Harris SF, Southworth DR, Agard DA (2006a) Structural analysis of E. coli Hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 127:329–340. doi:10.1016/j.cell.2006.09.027
Shiau AK, Harris SF, Southworth DR, Agard DA (2006b) Structural analysis of E. coli Hsp90 reveals dramatic nucleotide-dependent conformational rearrangements. Cell 127:329–340
Sievers F et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol. doi:10.1038/msb.2011.75
Southworth DR, Agard DA (2008) Species-dependent ensembles of conserved conformational states define the Hsp90 chaperone atpase cycle. Mol Cell 32:631–640. doi:10.1016/j.molcel.2008.10.024
Southworth DR, Agard DA (2011) Client-loading conformation of the Hsp90 molecular chaperone revealed in the cryo-em structure of the human Hsp90:Hop complex. Mol Cell 42:771–781. doi:10.1016/j.molcel.2011.04.023
Sprangers R, Kay LE (2007) Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature 445:618–622. doi:10.1038/nature05512
Stechmann A, Cavalier-Smith T (2004) Evolutionary origins of Hsp90 chaperones and a deep paralogy in their bacterial ancestors. J Eukaryot Microbiol 51:364–373
Street TO, Lavery LA, Verba KA, Lee CT, Mayer MP, Agard DA (2012) Cross-monomer substrate contacts reposition the Hsp90 N-terminal domain and prime the chaperone activity. J Mol Biol 415:3–15. doi:10.1016/j.jmb.2011.10.038
Tjandra N, Bax A (1997) Measurement of dipolar contributions to 1JCH splittings from magnetic-field dependence of J modulation in two-dimensional NMR spectra. J Magn Reson 124:512–515. doi:10.1006/jmre.1996.1088
Tugarinov V, Kay LE (2003) Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc 125:13868–13878. doi:10.1021/ja030345s
Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE (2003) Cross-correlated relaxation enhanced H-1-C-13 NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc 125:10420–10428. doi:10.1021/ja030153x
Tugarinov V, Kanelis V, Kay LE (2006) Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc 1:749–754. doi:10.1038/nprot.2006.101
Valafar H, Prestegard JH (2004) Redcat: a residual dipolar coupling analysis tool. J Magn Reson 167:228–241. doi:10.1016/j.jmr.2003.12.012
Verba KA, Wang RYR, Arakawa A, Liu YX, Shirouzu M, Yokoyama S, Agard DA (2016) Structural biology atomic structure of Hsp90-Cdc37-Cdk4 reveals that Hsp90 traps and stabilizes an unfolded kinase. Science 352:1542–1547. doi:10.1126/science.aaf5023
Xiao R et al (2010) The high-throughput protein sample production platform of the northeast structural genomics consortium. J Struct Biol 172:21–33. doi:10.1016/j.jsb.2010.07.011
Yagi H, Pilla KB, Maleckis A, Graham B, Huber T, Otting G (2013) Three dimensional protein fold determination from backbone amide pseudocontact shifts generated by lanthanide tags at multiple sites. Structure 21:883–890
Acknowledgements
We gratefully acknowledge financial support from the National Institute of General Medical Sciences (NIGMS Protein Structure Initiative grant U54GM094597 (GTM), biotechnology resource Grant P41GM103390 and Grant R01GM033225 (JHP)) and from the HHMI (DAA). Molecular graphics and structural analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311). Manuscript content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Additional information
Kari Pederson and Gordon R. Chalmers have contributed equally to the work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pederson, K., Chalmers, G.R., Gao, Q. et al. NMR characterization of HtpG, the E. coli Hsp90, using sparse labeling with 13C-methyl alanine. J Biomol NMR 68, 225–236 (2017). https://doi.org/10.1007/s10858-017-0123-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-017-0123-8