Abstract
Purpose
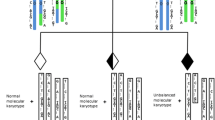
To explore a new preimplantation genetic testing (PGT) method for de novo mutations (DNMs) combined with chromosomal balanced translocations by whole-genome sequencing (WGS) using the MGISEQ-2000 sequencer.
Methods
Two families, one with maternal Olmsted syndrome caused by DNM (c.1246C>T) in TRPV3 and a paternal Robertsonian translocation and one with paternal Marfan syndrome caused by DNM (c.4952_4955delAATG) in FBN1 and a maternal reciprocal translocation, underwent PGT for monogenetic disease (PGT-M), chromosomal aneuploidy, and structural rearrangement. WGS of embryos and family members were performed. Bioinformatics analysis based on gradient sequencing depth was performed, and parent-embryo haplotyping was conducted for DNM diagnosis. Sanger sequencing, karyotyping, and chromosomal microarray analysis were performed using an amniotic fluid sample to confirm the PGT results.
Results
After 1 PGT cycle, WGS of 2 embryos from the Olmsted syndrome family revealed euploid embryos without DNMs; after 2 cycles, the 11 embryos from the Marfan syndrome family showed only 1 normal embryo without DNM, copy number variations (CNVs), or aneuploidy. Moreover, 1 blastocyst from the Marfan syndrome family was transferred back to the uterus; the amniocentesis test results were confirmed by PGT and a healthy infant was born.
Conclusions
WGS based on parent-embryo haplotypes was an effective strategy for PGT of DNMs combined with a chromosomal balanced translocation. Our results indicate this is a reliable and effective diagnostic method that is useful for clinical application in PGT of patients with DNMs.



Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–75.
Mao R, McDonald J, Cantwell M, Tang W, Ward K. The implication of de novo 21-hydroxylase mutation in clinical and prenatal molecular diagnoses. Genet Test. 2005;9:121–5.
Harton GL, De Rycke M, Fiorentino F, Moutou C, SenGupta S, Traeger-Synodinos J, et al. ESHRE PGD consortium best practice guidelines for amplification-based PGD. Hum Reprod. 2011;26:33–40.
Altarescu G, Eldar-Geva T, Varshower I, Brooks B, Haran EZ, Margalioth EJ, et al. Real-time reverse linkage using polar body analysis for preimplantation genetic diagnosis in female carriers of de novo mutations. Hum Reprod. 2009;24:3225–9.
Chen L, Diao Z, Xu Z, Zhou J, Yan G, Sun H. The clinical application of single-sperm-based SNP haplotyping for PGD of osteogenesis imperfecta. Syst Biol Reprod Med. 2019;65:75–80.
Rechitsky S, Pomerantseva E, Pakhalchuk T, Pauling D, Verlinsky O, Kuliev A. First systematic experience of preimplantation genetic diagnosis for de-novo mutations. Reprod BioMed Online. 2011;22:350–61.
Fiorentino F, Bono S, Biricik A, Nuccitelli A, Cotroneo E, Cottone G, et al. Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles. Hum Reprod. 2014;29:2802–13.
Handyside AH, Harton GL, Mariani B, Thornhill AR, Affara N, Shaw MA, et al. Karyomapping: a universal method for genome wide analysis of genetic disease based on mapping crossovers between parental haplotypes. J Med Genet. 2010;47:651–8.
Wang S, Niu Z, Wang H, Ma M, Zhang W, Fang Wang S, et al. De novo paternal FBN1 mutation detected in embryos before implantation. Med Sci Monit. 2017;23:3136–46.
Yan L, Huang L, Xu L, Huang J, Ma F, Zhu X, et al. Live births after simultaneous avoidance of monogenic diseases and chromosome abnormality by next-generation sequencing with linkage analyses. Proc Natl Acad Sci U S A. 2015;112:15964–9.
Gardner DK, Schoolcraft WB. In vitro culture of human blastocysts. In: Jansen R, Mortimer D, editors. Toward reproductive certainty: fertility and genetics beyond 1999. London: Parthenon Publishing; 1999. p. 378–88.
Gardner DK, Schoolcraft WB. Culture and transfer of human blastocysts. Curr Opin Obstet Gynecol. 1999;11:307–11.
Harton GL, Magli MC, Lundin K, Montag M, Lemmen J, Harper JC, et al. ESHRE PGD Consortium/Embryology Special Interest Group--best practice guidelines for polar body and embryo biopsy for preimplantation genetic diagnosis/screening (PGD/PGS). Hum Reprod. 2011;26:41–6.
den Dunnen JT, Dalgleish R, Maglott DR, Hart RK, Greenblatt MS, McGowan-Jordan J, et al. HGVS recommendations for the description of sequence variants: 2016 update. Hum Mutat. 2016;37:564–9.
Yin X, Tan K, Vajta G, Jiang H, Tan Y, et al. Massively parallel sequencing for chromosomal abnormality testing in trophectoderm cells of human blastocysts. Biol Reprod. 2013;88:69.
Giménez C, Sarasa J, Arjona C, Vilamajó E, Martínez-Pasarell O, Wheeler K, et al. Karyomapping allows preimplantation genetic diagnosis of a de-novo deletion undetectable using conventional PGD technology. Reprod BioMed Online. 2015;31:770–5.
Spits C, Le Caignec C, De Rycke M, Van Haute L, Van Steirteghem A, Liebaers I, et al. Whole-genome multiple displacement amplification from single cells. Nat Protoc. 2006;1:1965–70.
Hou Y, Song L, Zhu P, Zhang B, Tao Y, Xu X, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–85.
Peters BA, Kermani BG, Alferov O, Agarwal MR, McElwain MA, Gulbahce N, et al. Detection and phasing of single base de novo mutations in biopsies from human in vitro fertilized embryos by advanced whole-genome sequencing. Genome Res. 2015;25:426–34.
Masset H, Zamani Esteki M, Dimitriadou E, Dreesen J, Debrock S, Derhaag J, et al. Multi-Centre evaluation of a comprehensive preimplantation genetic test through haplotyping-by-sequencing. Hum Reprod. 2019;34:1608–19.
Acknowledgments
The authors thank all patients and their family members for their participation in this study.
Funding
This work was partially supported by the National Natural Science Foundation of China (No. 81801431), the Chinese Medical Association clinical medical research special fund, Research and development of young physicians in reproductive medicine (No. 18010060735), the Shenzhen Birth Defect Screening Project Lab (JZF No. [2016] 750), and the Shenzhen Municipal Government of China (No. JCYJ20170412152854656).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
Sanger sequencing results of the Olmsted syndrome family, their 2 embryos, and related the first polar body (PB1) and the second polar body (PB2) for detecting maternal missense mutation (c.1246C > T) in the TRPV3 gene. (PNG 134 kb)
Figure S2
Sanger sequencing results of the Marfan syndrome family and their 11 embryos for detecting paternal frameshift mutation (c.4952_4955delAATG) in the FBN1 gene. (PNG 221 kb)
Fig. S3
Validations of the fetus in the Marfan syndrome family. (a) Sanger sequencing of the amniotic fluid cells confirms that the baby does not have the mutant allele in TRPV3. (b) Chromosomal karyotype analysis reveals that the fetus has the same karyotype as the mother who carries the chromosomal reciprocal translocation (46, XN, t (10; 20) (q26.1; q13.1)). (c) Single nucleotide polymorphism (SNP) array for the detection of copy number variations (CNVs) and aneuploidy shows that the fetus has no chromosomal abnormalities. (PNG 3208 kb)
ESM 4
(DOCX 14 kb)
ESM 5
(DOCX 16 kb)
ESM 6
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Yuan, P., Xia, J., Ou, S. et al. A whole-genome sequencing–based novel preimplantation genetic testing method for de novo mutations combined with chromosomal balanced translocations. J Assist Reprod Genet 37, 2525–2533 (2020). https://doi.org/10.1007/s10815-020-01921-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-020-01921-4