Abstract
Turkey oak (Quercus cerris L.) is one of the most ecologically and economically significant deciduous tree species in the Central and Southeast European regions. The species has long been known to exhibit high levels of genetic and phenotypic variation. Recent climate response predictions for Turkey oak suggest a significant distribution extension in Europe under climate change. Since Turkey oak has relative drought-tolerant behaviour, it is regarded as a potential alternative for other forest tree species during forestry climate adaptation efforts, not only in its native regions but also in Western Europe. For this reason, the survey of existing genetic variability, genetic resources, and adaptability of this species has great significance. Next-generation sequencing approaches, such as ddRAD-seq (Double digest restriction-site associated DNA sequencing), allow the attainment of high-resolution genome-wide single nucleotide polymorphisms (SNPs). This study provides the first highly variable genome-wide SNP data for Turkey oak generated by ddRAD-seq. The dataset comprises 17 607 de novo and 26 059 reference mapped SNPs for 88 individuals from eight populations, two from Bulgaria, one from Kosovo, and five from Hungary. Reference mapping was carried out by using cork oak’s (Quercus suber L.) reference genome. The obtained high-resolution genome-wide markers are suitable for investigating selection and local adaptation and inferring genetic diversity, differentiation, and population structure. The dataset is accessible at: https://doi.org/10.5281/zenodo.8091252
Similar content being viewed by others
Introduction
The genetic architecture of oaks at different levels has been the subject of countless studies for a long time. Molecular genetic approaches are generally used to resolve taxonomic issues, infer phylogeny, population genetic structures, and adaptation of oaks (Conte et al. 2007; Simeone et al. 2016; Sork et al. 2016; Carabeo et al. 2017; Hipp et al. 2020). The intensive development of genetic approaches is opening new horizons in the study of oaks’ taxonomy and adaptive evolution. The advanced reduced representation approaches based on next-generation sequencing (e.g., RAD-seq) allow for the effective discovery of genome-wide single nucleotide polymorphisms (SNPs) obtaining high-resolution population genetic data, including non-model organisms such as forest trees (Davey and Blaxter 2010; Parchman et al. 2018). Through properly selected restriction enzyme or enzyme combinations, the restriction site-associated DNA sequencing (RAD-seq) method delivers thousands of SNP markers evenly covering the genome under study during genotypes of large numbers of individuals across many populations (Miller et al. 2007; Baird et al. 2008). Double digest restriction site-associated DNA sequencing (ddRAD-seq) is a two-restriction enzyme approach of RAD-seq, increasingly applied for the population genetic, phylogenetic, and candidate genes studies of oaks (Konar et al. 2017; López de Heredia et al. 2020; Tóth et al. 2021, 2023).
According to most recent predictions, climate change significantly affects the European climate. While the amplitude and duration of extreme events are increasing, forest site conditions get more arid characters (IPCC 2022). As part of the active adaptation efforts, forestry management is looking towards alternative tree species and their selected provenances that can cope with changing conditions, thereby increasing the biodiversity and resilience of forest ecosystems.
Turkey oak (Quercus cerris L.) is one of the most significant members of Quercus section Cerris, distributing from the Apennine Peninsula throughout the Balkans to Asia Minor. The north part of the area reaches the northern regions of the Carpathian basin, near 49° N; thus, Turkey oak is currently the northernmost penetrating species of the section and the whole subgenus Cerris (Old World oaks) (Menitsky 2005; Simeone et al. 2019). Recent dendrochronological studies suggest a differing drought adaptation strategy for Turkey oak and a higher tolerance against drought events than the most widespread white oak species (Quercus petraea (Matt.) Liebl.) in Hungary (Móricz et al. 2021; Mészáros et al. 2022). According to climate model predictions, Turkey oak habitats are expected to expand in response to climate change during this century (Thurm et al. 2018; Illés and Móricz 2022). Due to the drought-tolerant behaviour and the expected expansion, Turkey oak has great potential in future forestry adaptation efforts, not just in its native range but also in Western Europe. Therefore, deepening our knowledge of the species’ genetic makeup and adaptability is indispensable. According to the findings of available studies Turkey oak has a prominently high phenotypic and genetic variability with an increasing NW to SE gradient throughout its distribution range (Özer 2014; Bagnoli et al. 2016). However, currently, no high-resolution genome-wide level information is available for the species which would allow for revealing deep population structures and include functional genomic regions for association studies. The present study establishes a de novo and a reference-mapped genome-wide SNP dataset of eight Central and Southeast European Turkey oak populations to allow a detailed survey of the genetic variability and adaptability of the species in the investigated regions. We assume that through the thousands of SNP markers discovered and mapped to cork oak’s (Quercus suber L.) reference genome, gene flow, demography, and local adaptation processes of Turkey oak can be studied in more detail than ever before.
Our highly variable genome-wide SNP dataset through more than 17 K de novo and 26 K reference mapped SNPs allows deep insight into the genetic structure of eight Central and Southeast European Turkey oak populations for the first time. Combining reference-mapped SNPs with environmental conditions also provides opportunities for genome-wide association studies on the adaptation processes connected to changing climatic parameters and other related studies. In addition, this dataset was developed following the same methodology as the recently published SNP dataset of sessile oak (Tóth et al. 2021). The analyzed populations were collected from overlapping geographic regions; thus, there is a unique opportunity for comparative studies on the co-occurring white oak and a Cerris oak species.
Material and methods
Plant material
During the sampling work, we collected plant material from five populations in Hungary, two in Bulgaria, and one from Kosovo. The broader sampling regions were carefully selected in relation to findings concerning biogeography, the existing spatial genetic structure of the species under study, and the co-occurring white oaks (Bordács et al. 2002; Petit et al. 2002; Özer 2014; Bagnoli et al. 2016; Gömöry et al. 2020). To ensure complex evaluability and comparability, the exact sampling locations were selected from the sessile oak sampling sites in Tóth et al. (2021), in the Balkans, from within the Turkey oak – sessile oak dendrochronology sites in Móricz et al. (2021), and from the experimental site of the Síkfőkút Project in Hungary (Mészáros et al. 2022). Finally, a total of eight even-aged Turkey oak stands with assumed local origin were selected, two from Bulgaria (Balgari and Botevgrad), one from Kosovo (Pristina), and five from Hungary (Zselickisfalud, Iharos, Vállus, Szántód and Síkfőkút), respectively (Fig. 1, Table 1).
Geographic distribution of sampling locations (the distribution range of Turkey oak in light green (Caudullo et al. 2022))
During the sampling process, we collected fresh leaves from 88 mature trees from the eight populations and stored them by freezing (− 20 °C) or drying them with silica gel until processing. The spatial coordinates (GPS) of the population in every sampling location were recorded and a minimum of 30 m isolation distance was maintained to avoid sampling-related individuals.
Library preparation and ddRAD-sequencing
During lab processing, the total genomic DNA was extracted from seven leaf disks (5 mm in diameter) per sampled individual following the method of Dumolin et al. (1995), except for the HU3 population where the QIAGEN DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany) was used. Next, extracted DNA samples were quantified by a Qubit 3.0 Fluorometer using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). For the double-digestion process of the DNA samples, the combination of Pst I and Msp I restriction enzymes was selected according to the findings of Cumer et al. (2021). In advance restriction sites were analyzed in the CLC Genomic Workbench software version 12.0 (QIAGEN Bioinformatics, Hilden, Germany). Double-digestion was performed on 50 ng of DNA per sample at 37 °C for two hours using 0.1 µl of each enzyme Pst I and Msp I respectively (FastDigest restriction enzymes; Thermo Fisher Scientific, Waltham, MA, USA). Fragments were double-sided size selected by KAPA PureBeads with a 0.55–0.80X solution/bead ratio (Roche, Basel, Switzerland) to isolate 300–600 bp fragments of digested DNA. Then the selected fragments were ligated to oligonucleotide adapters (Table 2) using T4 DNA Ligase, according to the producer protocol (Thermo Fisher Scientific, Waltham, MA, USA). Ligated fragments were purified using 0.8 KAPA PureBeads (Roche, Basel, Switzerland), and then amplified PCR using NEBNext Oligos for Illumina (Dual Index Set 1; New England Biolabs, Ipswich, MA, USA) and KAPA HiFi Hotstart Ready Mix (Roche, Basel, Switzerland). During each amplification reaction, i5 and i7 indexed primers were used in the same 0.5 µl amount. The thermal conditions of the PCR cycles were set in the following order: initial denaturation for three minutes at 95 °C, denaturation for 30 s at 95 °C in 17 repetitions, annealing for 30 s at 55 °C, extension for 30 s at 72 °C, and a final extension for five minutes at 72 °C. After amplification, the quantity and quality of the created amplicon library were checked using a High Sensitivity DNA1000 ScreenTape system with 2200 Tapestation (Agilent Technologies, Santa Clara, CA, USA) and dsDNA HS Assay Kit with Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). Before the paired-end (2 × 300-bp) sequencing, the equimolarily pooled libraries were diluted to 10 pM using the 600-cycle sequencing kit v3.1 (Illumina, San Diego, CA, USA). Finally, next-generation sequencing of the prepared libraries was performed on a MiSeq Sequencing System (Illumina, San Diego, CA, USA) according to the producer protocol. Raw sequence data were uploaded to the NCBI Sequence Read Archive (SRA) and are available under the BioProject ID: PRJNA927246.
Bioinformatic processes of raw sequences
All bioinformatics processing work was performed on the Silicon Computers (SGI) HPC server of the University of Sopron Forest Research Institute (Sopron, Hungary) with 40 cores (80 threads) and 38 GB RAM. In the first step using MiSeq Control Software (Illumina, San Diego, CA, USA), raw reads of Illumina sequencing were demultiplexed, and then the adapter sequences from the ends of reads were trimmed. In the next step, sequences at the 3´ and 5´ end with a quality score lower than 30 were trimmed. Reads with a mean quality score lower than 30 or a length shorter than 200 bp were also filtered out by using FastQ Toolkit. The purified raw data were further computationally processed using Stacks 2.0 software (Catchen et al. 2013; Rochette et al. 2019). This processing phase was initiated with additional filtering and trimming on the 37 701 348 raw sequences retained in the previous step, implemented in the program “process_radtags”. With the sliding-window method (the 15% of the given read length), every read that has a quality score lower than 90% (raw Phred score of 10) was filtered out (Catchen et al. 2011). In addition, to allow proper assembly and omit the low-quality sequences at the end of reads, all reads were uniformly truncated to 200 bp (Catchen et al. 2011). After filtering out 384 290 low-quality and 6 143 450 adapter-containing reads, a total of 31 173 608 sequences were retained for further analysis. Thus, purified and uniformed reads were now suitable to the de novo assembly of RAD loci and the SNP calling.
De novo assembly and reference mapping
To allow multipurpose usage of our dataset we performed both de novo and reference mapped assembly. For de novo variant calling we implemented the de novo pipeline using the function “denovo_map.pl” of STACKS. This program is composed of three successive functions, “ustacks”, “cstacks” and “sstacks”, respectively (Catchen et al. 2011; Rochette et al. 2019). Before running the de novo pipeline, command parameters had to be optimized for the given dataset. These required parameters are M, the maximum number of mismatches allowed between two alleles; m, the minimum number of reads to form an allele; n, the maximum number of mismatches permitted between two individual loci to consider them as homologous (Mastretta-Yanes et al. 2015; Paris et al. 2017). Optimization was performed following the “r80” method, which maximizes the number of SNPs found in 80% of the individuals (Paris et al. 2017). To find the optimal M and n values, nine test runs of denovo_map.pl were done following the M = n rule in each run by setting its values from two to ten. The optimal value of M and n parameters is the one after which increasing the parameters no longer leads to the detection of more new SNPs (Fig. S1a). According to this approach, the value of M and n parameters was chosen to be three (Paris et al. 2017). For the value of the third parameter m the minimum number of reads to form an allele, the default value, 3 was selected. In the next step, a final run was executed with the above-selected parameters. After using the command “gstacks”, paired-end contigs were assembled, and SNPs were recalled (Rochette et al. 2019).
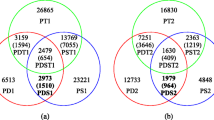
As a result, assembly and alignment of paired-end reads were conducted for 57 285 loci, from which 559 had paired-end reads that could not be assembled into a contig (1.0%). The assembly of a paired-end contig was successful for the remaining 56 726 loci (99.0%), with an average contig size of 319.3 bp. The mean of effective per-sample coverage was given as 16.1 × with 2.7 × standard deviation, 7.0 × minimum and 30.2 × maximum values. Table E S1 summarizes the per-population values; for a graphical visualization, see Fig. SI1b.
Reference mapping was carried out on the cork oak reference genome ( GCA_002906115.4 [haploid version]; https://www.corkoakdb.org; Ramos et al. 2018), as this species is the closest relative of Turkey oak among the complete genome sequenced oak species. To map our raw reads against the reference genome, we used the BWA-MEM v0.7.17 software (Li 2013), previously proving its effectiveness during the reference mapping of several oak species (e.g. Tóth et al. (2023)). During the mapping process, we used the software’s default parameters and discarded the unassigned scaffolds of the reference genome. After mapping, we sorted the resulting SAM files containing the sequences of individual samples by using SAMtools v1.10 software. In the next step, SAM files were converted to BAM files, and then reads with a mapping quality lower than five (MAPQ > = 5) were removed from the dataset. Finally, we calculated individual- and population-level summary statistics by using ‘flagstat’ function in SAMtools (Fig. 2, Table S1, Table S1).
After reference mapping, variant calling was also carried out in STACKS software by using ‘qstacks’ program. During this, loci were reconstructed and a SNP catalogue was created by using sliding window algorithm. In addition, unpaired reads which would support SNPs aligned to only one strand (strand-bias error) were removed.
The final step of the variant calling process was the same for both the de novo and the reference-mapped datasets. This final filtering was implemented in the “populations” program of STACKS and its goal was to discard insecure SNPs (Catchen et al. 2011). During this, every SNP loci with a minor allele frequency lower than 0.05, a rate of missing individuals greater than 0.2 or a significant deviation from Hardy–Weinberg equilibrium (HWE, P < 0.001) was removed from the datasets (Catchen et al. 2011). In addition, the “minimum number of populations” value was set to default 1, which means that SNPs that occurred in only one population were also retained. This allows future users to further filter the datasets according to their study requirements.
For the de novo dataset, after implementing the filtering thresholds, 51 861 of the 56 725 loci did not pass the sample/population, and minor allele frequency constraints and 4206 sites were discarded because they were significantly outside the Hardy–Weinberg equilibrium. The final de novo dataset, thus, comprises 1 662 692 sites in total, containing 17 607 variant sites (SNPs) at 4865 loci.
In some cases, reference mapping could result in greater efficiency of loci discovery (Fitz-Gibbon et al. 2017; Shafer et al. 2017; Ulaszewski et al. 2021; Vaux et al. 2023). During our reference mapping, we also achieved higher success in loci assembly than de novo pipeline. As a result, we obtained 7184 loci with 2 908 293 sites, of which 53 923 were filtered out, and 26 059 variant sites were retained. This means a roughly 50% increase in the number of discovered SNPs.
Access dataset
The final SNP dataset was deposited at the ZENODO repository and is available via the following link: https://doi.org/10.5281/zenodo.8091252. Both de novo and reference mapped datasets inside the repository are in two PLINK formatted, UTF-8 encoded, tab-delimited ped, and map files. By using the freely available genomic analyzing software PLINK, the uploaded dataset can be easily manipulated, converted to several other genomic formats, or directly utilized in genetic tests (Purcell et al. 2007). The first column inside the ped extension file comprises the population IDs from 1 to 8, while the second column contains the individual IDs. The data in the third to sixth columns is undefined (mother ID, father ID, sex, and phenotype data). Data in the seventh column onwards include nucleotide sequences (coded as A, T, G, C), where every pair of two columns correspond to a diploid SNP locus. Missing data were coded as “0 0” according to standard PLINK formatting (see https://zzz.bwh.harvard.edu/plink/). In the first column of the map file, the number of chromosomes was coded to undefined in the de novo dataset. As for the reference mapped dataset, the first column of the map file contains the scaffold ID where the given SNP is located since cork oak’s genome was assembled only into scaffolds, not chromosomes. The second column contains the SNP IDs in the case of both de novo and reference-mapped files. The third column has non-defined values in both cases (genetic position of a given SNP), and the fourth column includes the physical positions of SNPs on RAD loci or the given scaffold by reference mapped data in bp. Elevation and GPS coordinates are available for every population in Table 1.
Quality control of raw sequences
The quality check of raw short-read data for every individual was performed using the FastQC v0.11.9 program (Andrews 2010) in three successive steps. In the first step, per-base and per-sequence quality was checked directly after sequencing; in the second step, after trimming bad quality bases from the 3´ and 5´ end; and in the third step, at sequence processing (whole read filtering). In each step, the 176 produced reports were combined and collectively evaluated with the MultiQC v1.14 program (Ewels et al. 2016), which allows for the effective simultaneous evaluation of large numbers of FastQC reports (Fig. SI2). Since the raw data was consistent with standard Illumina run in every quality control step, its quality was high enough for further processing (Kircher et al. 2011).
Applications
Although reduced representation technologies (such as RAD-seq) are becoming widely available, high-resolution datasets are still considered rare, particularly for forest tree species without reference genomes. The number of studies based on genome-wide SNP data is increasing in oak genetics, though mainly for white oaks. This dataset delivers highly polymorphic SNP markers for Turkey oak belonging to the group of Old World oaks for the first time. In addition, the sampling area affected the genetically highly diverse but scarcely studied Pannonian and Balkan regions. Since this dataset was generated in the same way as the recently published SNP dataset of sessile oak (Tóth et al. 2021) covering the same geographic regions, there is a unique opportunity for the comparative study of the two genetically relatively distant oak species.
For a preliminary population genetic overview basic descriptive indices were computed by using packages “adegenet” (Jombart and Ahmed 2011), “hierfstat” (Goudet and Jombart 2022), and “poppr” (Kamvar et al. 2014) in the statistical software R (R Core Team 2022). Table 3 summarizes the within-population values of observed heterozygosity (Ho), gene diversity (Hs), and the inbreeding coefficient (Fis) following Nei (1987). The calculated indices were compared with the same values from the sessile oak dataset (Tóth et al. 2021), as no such data are currently available for Turkey oak. According to the comparison, indices of Turkey oak populations were quite similar to the values of sessile oak populations except for the Fis values, which are higher in this case. According to the calculated 95% confidence intervals for mean Fis, the inbreeding coefficient is significantly greater than zero in each population. However, note only preliminary filtering was carried out on this dataset to ensure more flexible use. Accordingly, careful filtering may needed before population genetic analysis in line with the objectives of the given study.
Using the more than 17 K de novo and 26 K reference mapped highly polymorphic SNP markers creates several opportunities to study population genetic and genomic aspects of Turkey oak. The gene flow and spatial genetic structure of populations can be investigated in depth by considering geographic locations. In addition, high-resolution SNPs enable the fine-scale study of kinship networks inside populations and hybridization. On a larger temporal scale, discovering phylogeography and mapping glacial refugia and postglacial migration routes tracking the consequences of ancient hybridization and population migration (contraction and re-expansion) events become possible. Combining the reference-mapped dataset with climatic data also gives opportunities for genome-wide association studies.
Nevertheless, we draw the attention of future users that, as with every reduced representation method generated dataset, population genetic inference taken from this dataset should also be treated with particular care since these methods (such as ddRAD-seq) often introduce biases (Arnold et al. 2013; Cariou et al. 2016). Although RAD-seq generated data may have more adverse attributes concerning phylogeny estimation (short loci, low coverage and SNP density, non-random sampling because of the polymorphisms of restriction sites), with appropriate data care, the method is suitable for phylogenetic studies as well (Cariou et al. 2013; Hühn et al. 2022).
Data availability
The SNP datasets generated during the current study are available in the ZENODO repository, https://doi.org/10.5281/zenodo.8091252. Raw sequences are available on NCBI SRA database under the BioProject ID: PRJNA927246 [accession numbers: SAMN32903873—SAMN32903960].
References
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom
Arnold B, Corbett-Detig RB, Hartl D, Bomblies K (2013) RADseq underestimates diversity and introduces genealogical biases due to nonrandom haplotype sampling. Mol Ecol 22:3179–3190. https://doi.org/10.1111/mec.12276
Bagnoli F, Tsuda Y, Fineschi S et al (2016) Combining molecular and fossil data to infer demographic history of Quercus cerris: Insights on European eastern glacial refugia. J Biogeogr 43:679–690. https://doi.org/10.1111/jbi.12673
Baird NA, Etter PD, Atwood TS et al (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376. https://doi.org/10.1371/journal.pone.0003376
Bordács S, Popescu F, Slade D et al (2002) Chloroplast DNA variation of white oaks in northern Balkans and in the Carpathian Basin. For Ecol Manage 156:197–209. https://doi.org/10.1016/S0378-1127(01)00643-0
Carabeo M, Simeone MC, Cherubini M et al (2017) Estimating the genetic diversity and structure of Quercus trojana webb populations in Italy by SSRs: implications for management and conservation. Can J for Res 47:331–339. https://doi.org/10.1139/cjfr-2016-0311
Cariou M, Duret L, Charlat S (2013) Is RAD-seq suitable for phylogenetic inference? An in silico assessment and optimization. Ecol Evol 3:846–852. https://doi.org/10.1002/ece3.512
Cariou M, Duret L, Charlat S (2016) How and how much does RAD-seq bias genetic diversity estimates? BMC Evol Biol 16:240. https://doi.org/10.1186/s12862-016-0791-0
Catchen JM, Amores A, Hohenlohe P et al (2011) Stacks: building and genotyping Loci De Novo from short-read sequences. G3 Genes|genomes|genetics 1:171–182. https://doi.org/10.1534/g3.111.000240
Catchen JM, Hohenlohe PA, Bassham S et al (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140. https://doi.org/10.1111/mec.12354
Caudullo G, Welk E, San-Miguel-Ayanz J (2022) Chorological data for the main European woody species. Mendeley Data. https://doi.org/10.17632/hr5h2hcgg4.15
Conte L, Cotti C, Cristofolini G (2007) Molecular evidence for hybrid origin of Quercus crenata Lam. (Fagaceae) from Q. cerris L. and Q. suber L. Plant Biosyst - an Int J Deal with All Asp Plant Biol 141:181–193. https://doi.org/10.1080/11263500701401463
Cumer T, Pouchon C, Boyer F et al (2021) Double-digest RAD-sequencing: do pre- and post-sequencing protocol parameters impact biological results? Mol Genet Genomics 296:457–471. https://doi.org/10.1007/s00438-020-01756-9
Davey JW, Blaxter ML (2010) RADSeq: next-generation population genetics. Brief Funct Genomics 9:416–423. https://doi.org/10.1093/bfgp/elq031
Dumolin S, Demesure B, Petit RJ (1995) Inheritance of chloroplast and mitochondrial genomes in pedunculate oak investigated with an efficient PCR method. Theor Appl Genet 91:1253–1256. https://doi.org/10.1007/BF00220937
Ewels P, Magnusson M, Lundin S, Käller M (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. https://doi.org/10.1093/bioinformatics/btw354
Fitz-Gibbon S, Hipp AL, Pham KK et al (2017) Phylogenomic inferences from reference-mapped and de novo assembled short-read sequence data using RADseq sequencing of California white oaks (Quercus section Quercus). Genome 60:743–755. https://doi.org/10.1139/gen-2016-0202
Gömöry D, Zhelev P, Brus R (2020) The Balkans: a genetic hotspot but not a universal colonization source for trees. Plant Syst Evol 306:5. https://doi.org/10.1007/s00606-020-01647-x
Goudet J, Jombart T (2022) hierfstat: Estimation and Tests of Hierarchical F-Statistics. https://cran.r-project.org/package=hierfstat
Hipp AL, Manos PS, Hahn M et al (2020) Genomic landscape of the global oak phylogeny. New Phytol 226:1198–1212. https://doi.org/10.1111/NPH.16162
Hühn P, Dillenberger MS, Gerschwitz-Eidt M et al (2022) How challenging RADseq data turned out to favor coalescent-based species tree inference A case study in Aichryson (Crassulaceae). Mol Phylogenet Evol 167:107342. https://doi.org/10.1016/j.ympev.2021.107342
Illés G, Móricz N (2022) Climate envelope analyses suggests significant rearrangements in the distribution ranges of Central European tree species. Ann for Sci 79:35. https://doi.org/10.1186/s13595-022-01154-8
IPCC (2022) Climate Change 2022: Impacts Adaptation and Vulnerability. Cambridge University Press, Cambridge, UK and New York, NY, USA
Jombart T, Ahmed I (2011) adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27:3070–3071. https://doi.org/10.1093/bioinformatics/btr521
Kamvar ZN, Tabima JF, Grünwald NJ (2014) Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281. https://doi.org/10.7717/peerj.281
Kircher M, Heyn P, Kelso J (2011) Addressing challenges in the production and analysis of illumina sequencing data. BMC Genomics 12:382. https://doi.org/10.1186/1471-2164-12-382
Konar A, Choudhury O, Bullis R et al (2017) High-quality genetic mapping with ddRADseq in the non-model tree Quercus rubra. BMC Genomics 18:417. https://doi.org/10.1186/s12864-017-3765-8
Li H (2013) Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv: Genomics. https://doi.org/10.48550/arXiv.1303.3997
López de Heredia U, Mora-Márquez F, Goicoechea PG et al (2020) ddRAD sequencing-based identification of genomic boundaries and permeability in Quercus ilex and Q suber hybrids. Front Plant Sci 11:564414. https://doi.org/10.3389/fpls.2020.564414
Mastretta-Yanes A, Arrigo N, Alvarez N et al (2015) Restriction site-associated DNA sequencing, genotyping error estimation and de novo assembly optimization for population genetic inference. Mol Ecol Resour 15:28–41. https://doi.org/10.1111/1755-0998.12291
Menitsky YL (2005) Oaks of Asia, Science Publishes Inc., Enfield, New Hampshire, USA. Translated from Russian
Mészáros I, Adorján B, Nyitrai B et al (2022) Long-term radial growth and climate-growth relationships of Quercus petraea (Matt) Liebl. and Quercus cerris L. in a xeric low elevation site from Hungary. Dendrochronologia 76:126014. https://doi.org/10.1016/j.dendro.2022.126014
Miller MR, Dunham JP, Amores A et al (2007) Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 17:240–248. https://doi.org/10.1101/gr.5681207
Móricz N, Illés G, Mészáros I et al (2021) Different drought sensitivity traits of young sessile oak (Quercus petraea (Matt.) Liebl.) and Turkey oak (Quercus cerris L.) stands along a precipitation gradient in Hungary. For Ecol Manage 492:119165. https://doi.org/10.1016/j.foreco.2021.119165
Nei M (1987) Molecular Evolutionary Genetics Columbia University Press. New York Chichester, West Sussex
Özer TY (2014) Patterns of Genetic Diversity in Turkey Oak (Quercus cerris L.) Populations. Dissertation, Middle East Technical University, Ankara, Turkey. Retrieved from: https://open.metu.edu.tr/handle/11511/23662
Parchman TL, Jahner JP, Uckele KA et al (2018) (2018) RADseq approaches and applications for forest tree genetics. Tree Genet Genomes 143(14):1–25. https://doi.org/10.1007/S11295-018-1251-3
Paris JR, Stevens JR, Catchen JM (2017) Lost in parameter space: a road map for stacks. Methods Ecol Evol 8:1360–1373. https://doi.org/10.1111/2041-210X.12775
Petit RJ, Brewer S, Bordács S et al (2002) Identification of refugia and post-glacial colonisation routes of European white oaks based on chloroplast DNA and fossil pollen evidence. For Ecol Manage 156:49–74. https://doi.org/10.1016/S0378-1127(01)00634-X
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
R Core Team (2022) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http//www R-project org/
Ramos AM, Usié A, Barbosa P et al (2018) The draft genome sequence of cork oak. Sci Data 5:180069. https://doi.org/10.1038/sdata.2018.69
Rochette NC, Rivera-Colón AG, Catchen JM (2019) Stacks 2: Analytical methods for paired-end sequencing improve RADseq-based population genomics. Mol Ecol 28:4737–4754. https://doi.org/10.1111/mec.15253
Shafer ABA, Peart CR, Tusso S et al (2017) Bioinformatic processing of RAD-seq data dramatically impacts downstream population genetic inference. Methods Ecol Evol 8:907–917. https://doi.org/10.1111/2041-210X.12700
Simeone MC, Grimm GW, Papini A et al (2016) Plastome data reveal multiple geographic origins of Quercus Group Ilex. PeerJ. https://doi.org/10.7717/peerj.1897
Simeone MC, Stojanov PZ, Kandemir G (2019) EUFORGEN Technical Guidelines for genetic conservation and use of Turkey oak (Quercus cerris). Eur For Genet Resour Program (EUFORGEN), Eur For Inst 6
Sork VL, Squire K, Gugger PF et al (2016) Landscape genomic analysis of candidate genes for climate adaptation in a California endemic oak, Quercus lobata. Am J Bot 103:33–46. https://doi.org/10.3732/ajb.1500162
Thurm EA, Hernandez L, Baltensweiler A et al (2018) Alternative tree species under climate warming in managed European forests. For Ecol Manage 430:485–497. https://doi.org/10.1016/j.foreco.2018.08.028
Tóth EG, Köbölkuti ZA, Cseke K et al (2021) A genomic dataset of single-nucleotide polymorphisms generated by ddRAD tag sequencing in Q petraea (Matt.) Liebl. populations from Central-Eastern Europe and Balkan Peninsula. Ann for Sci 78:43. https://doi.org/10.1007/s13595-021-01051-6
Tóth EG, Cseke K, Benke A et al (2023) Key triggers of adaptive genetic variability of sessile oak [Q. petraea (Matt.) Liebl.] from the Balkan refugia: outlier detection and association of SNP loci from ddRAD-seq data. Heredity (edinb). https://doi.org/10.1038/s41437-023-00629-2
Ulaszewski B, Meger J, Burczyk J (2021) Comparative analysis of SNP discovery and genotyping in Fagus sylvatica L. and Quercus robur L. using RADseq GBS and ddRAD methods. Forests 12:222
Vaux F, Dutoit L, Fraser CI, Waters JM (2023) Genotyping-by-sequencing for biogeography. J Biogeogr 50:262–281. https://doi.org/10.1111/jbi.14516
Acknowledgements
The authors thank all those who contributed to the planning and implementation of the sample collection: Ivan Iliev, Petar Zhelev, and Vladimir T. Tomov (University of Forestry, Bulgaria), Ibrahim Muja (Ministry of Agriculture Forestry and Rural Development, (MAFRD), Kosovo), and Naser Krasniqi (Kosovo Forest Agency, Kosovo). Without their work, our study would not have been possible. We also thank Zoltan Bihari (Xenovea Ltd.) for providing the sequencing resources and useful technical advice on data processing.
Funding
Open access funding provided by University of Sopron. The sampling collection was founded by the Ministry of Agriculture of Hungary (Kaán Károly project, no.: EVgF/549/2018, EGF/178/2019). Bioinformatic processing was performed in the frame of the project TKP2021-NKTA-43 which has been implemented with the support provided by the Ministry of Innovation and Technology of Hungary (successor: Ministry of Culture and Innovation of Hungary) from the National Research, Development and Innovation Fund, financed under the TKP2021-NKTA funding scheme.
Author information
Authors and Affiliations
Contributions
BB.L, KC, and EG.T conceived the study. EG. T, ZA.K, KC, AB, NM, and TM.N contributed to the sampling of plant materials. KC performed laboratory analysis. BB.L and EG.T processed genomic data and performed statistical analyses and simulations. BB.L wrote the manuscript. CÉ.M, LN, AB, and IM provided suggestions for data analyses and feedback on manuscript drafts. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lados, B.B., Cseke, K., Benke, A. et al. ddRAD-seq generated genomic SNP dataset of Central and Southeast European Turkey oak (Quercus cerris L.) populations. Genet Resour Crop Evol (2024). https://doi.org/10.1007/s10722-024-01889-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10722-024-01889-5