Abstract
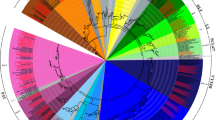
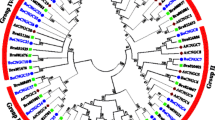
COBRA-like genes play important roles in oriented cell division, cell expansion, cell wall biosynthesis, defense response, and tip-directed growth in root hair development in plants. However, despite the increasing availability of plant genomic data, there is limited understanding of the systemic evolutionary analysis and gene characterization of this family. This study aims to investigate the number and structure of the COBRA gene family in land plants, representing major evolutionary nodes, through genome data mining. The representative plants analyzed in this study include Arabidopsis thaliana, Arabidopsis lyrate, Carica papaya, Populus trichocarpa, Medicago truncatula, Glycine max, Ricinus communis, Manihot esculenta, Cucumis sativus, Vitis vinifera, Sorghum bicolor, Zea mays, Oryza sativa, Brachypodium distachyon, Mimulus guttatus, Selaginella moellendorffii, and Phycomitrella patens. A total of 180 COBRA proteins were analyzed from these representative plants. By comparing protein and gene structures, combined with phylogenetic analysis, syntenic research, and expression pattern analysis, this study provides novel insights into the origin and divergence of COBRA genes. Furthermore, this study reveals the evolutionary relationships of COBRA genes among land plant species and offers essential theoretical foundations and gene resources for improving crop quality through genetic modification mechanisms.





Similar content being viewed by others
Explore related subjects
Discover the latest articles and news from researchers in related subjects, suggested using machine learning.References
Appella E, Weber IT, Blasi F (1988) Structure and function of epidermal growth factor-like regions in proteins. FEBS Lett 231:1–4
Ben-Tov D, Abraham Y, Stav S, Thompson K, Loraine A, Elbaum R, de Souza A, Pauly M, Kieber JJ, Harpaz-Saad S (2015) COBRA-LIKE2, a member of the glycosylphosphatidylinositol-anchored COBRA-LIKE family, plays a role in cellulose deposition in Arabidopsis seed coat mucilage secretory cells. Plant Physiol 167:711–724
Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119:57–70
Berardini BZ, Reiser L, Li D (2015) The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis 53:474–485
Brady SM, Song S, Dhugga KS, Rafalski JA, Benfey PN (2007a) Combining expression and comparative evolutionary analysis: The COBRA Gene Family. Plant Physiol 143:172–187
Bulmer M (1991) The selection-mutation-drift theory of synonymous codon usage. Genetics 129:897–907
Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Ching A, Dhugga KS, Appenzeller L, Meeley R, Bourett TM, Howard RJ, Rafalski A (2006) Brittle stalk 2 encodes a putative glycosylphosphatidylinositol-anchored protein that affects mechanical strength of maize tissues by altering the composition and structure of secondary cell walls. Planta 224:1174–1184
Csürös M, Holey JA, Rogozin IB (2007) In search of lost introns. Bioinformatics 23:87–96
Csürös M, Rogozin IB, Koonin EV (2008) Extremely intron-rich genes in the alveolate ancestors inferred with a flexible maximum-likelihood approach. Mol Biol Evol 25:903–911
Doolittle R, Fei Feng D, Johnson M (1984) Computer-based characterization of epidermal growth factor precursor. Nature 307:558–560
Eisenhaber B, Bork P, Eisenhaber F (1998) Sequence properties of GPI-anchored proteins near the omega-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng Des Sel 11:1155–1161
Emanuelsson O, Brunak S, Von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2:953–971
Goldman N, Yang ZH (1994) Codon-based model of nucleotide substitution for protein-coding dna-sequences. Mol Biol Evol 11:725–736
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hochholdinger F, Wen TJ, Zimmermann R, Chimot‐Marolle P, Da Costa e Silva O, Bruce W, Lamkey KR, Wienand U, Schnable PS (2008) The maize (Zea mays L.) roothairless3 gene encodes a putative GPI‐anchored, monocot‐specific, COBRA‐like protein that significantly affects grain yield. Plant J 54:888–898
Jones DT, Taylor WR, Thornton JM (1994) A mutation data matrix for transmembrane proteins. FEBS Lett 339:269–275
Kyte J, Doolittle R (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Li Y, Qian Q, Zhou Y, Yan M, Sun L, Zhang M, Fu Z, Wang Y, Han B, Pang X, Chen M (2003) BRITTLE CULM1, which encodes a COBRA-like protein, affects the mechanical properties of rice plants. Plant Cell 15:2020–2031
Liu L, Shang-Guan K, Zhang B, Liu X, Yan M, Zhang L, Shi Y, Zhang M, Qian Q, Li J, Zhou Y (2013) Brittle Culm1, a COBRA-like protein, functions in cellulose assembly through binding cellulose microfibrils. PLoS Genet 9:1003704
Lynch M (2002) Intron evolution as a population-genetic process. Proc Natl Acad Sci USA 99:6118–6123
Lynch M (2011) The lower bound to the evolution of mutation rates. Genome Biol Evol 3:1107–1118
McVean G, Charlesworth B (1999) A population genetic model for the evolution of synonymous codon usage: Patterns and predictions. Genetics Research 74:145–158
Reese MG (2001) Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem 26:51–56
Roudier F, Schindelman G, DeSalle R, Benfey PN (2002) The COBRA family of putative GPI-anchored proteins in Arabidopsis. a new fellowship in expansion. Plant Physiol 130:538–548
Schindelman G, Morikami A, Jung J, Baskin TI, Carpita NC, Derbyshire P, McCann MC, Benfey PN (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev 15:1115–1127
Shoemaker RC, Polzin KAYLA, Labate J, Specht JAMES, Brummer EC, Olson TERRY, Young NEVIN, Concibido V, Wilcox J, Tamulonis JP, Kochert G (1996) Genome duplication in soybean (Glycine subgenus soja). Genetics 144:329–338
Sindhu A, Langewisch T, Olek A, Multani DS, McCann MC, Vermerris W, Carpita NC, Johal G (2007) Maize Brittle stalk2 encodes a COBRA-like protein expressed in early organ development but required for tissue flexibility at maturity. Plant Physiol 145:1444–1459
Steel M (1994) Recovering a tree from the leaf colourations it generates under a Markov model. Appl Math Lett 7:19–23
Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M (2012) Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci USA 109:18488–18492
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson J, Higgins D, Gibson T (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Trivers R, Burt A, Palestis BG (2004) B chromosomes and genome size in flowering plants. Genome 47:1–8
Vinogradov AE (2001) Mirrored genome size distributions in monocot and dicot plants. Acta Biotheor 49:43–51
Yang ZH (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591
Ye X, Kang BG, Osburn LD, Cheng ZM (2009) The COBRA gene family in populus and gene expression in vegetative organs and in response to hormones and environmental stresses. Plant Growth Regul 58:211–223
Acknowledgments
Authors are thankful to the Researchers Supporting Project number (RSPD2023R728), King Saud University, Riyadh, Saudi Arabia.
Funding
Authors are thankful to the Researchers Supporting Project number (RSPD2023R728), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
M.Z. A, conducted the experimental work, performed the whole-genome analysis, and led the manuscript. A.S.A. and F.A.N. contributed to analysis gene expression data, and S.A.A contributed to editing, proofreading, and table arrangements and analysis.
Corresponding author
Ethics declarations
Competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ahmed, M.Z., Alqahtani, A.S., Nasr, F.A. et al. Comprehensive analysis of the COBRA-like (COBL) gene family through whole-genome analysis of land plants. Genet Resour Crop Evol 71, 863–872 (2024). https://doi.org/10.1007/s10722-023-01667-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-023-01667-9