Abstract
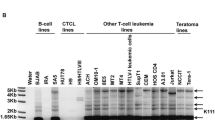
Transposable elements (TEs) account for nearly half (44 %) of the human genome. However, their overall activity has been steadily declining over the past 35–50 million years, so that <0.05 % of TEs are presumably still “alive” (potentially transposable) in human populations. All the active elements are retrotransposons, either autonomous (LINE-1 and possibly the endogenous retrovirus ERVK), or non-autonomous (Alu and SVA, whose transposition is dependent on the LINE-1 enzymatic machinery). Here we show that a lineage of the endogenous retrovirus ERVE was recently engaged in ectopic recombination events and may have at least one potentially fully functional representative, initially reported as a novel retrovirus isolated from blood cells of a Chinese patient with chronic myeloid leukemia, which bears signals of positive selection on its envelope region. Altogether, there is strong evidence that ERVE should be included in the short list of potentially active TEs, and we give clues on how to identify human specific insertions of this element that are likely to be segregating in some of our populations.



Similar content being viewed by others
References
Arnaud F, Caporale M, Varela M, Biek R, Chessa B et al (2007) A paradigm for virus–host coevolution: sequential counter-adaptations between endogenous and exogenous retroviruses. PLoS Pathog 3:e170
Balada E, Ordi-Ros J, Vilardell-Tarrés M (2009) Molecular mechanisms mediated by human endogenous retroviruses (HERVs) in autoimmunity. Rev Med Virol 19:273–286
Bannert N, Kurth R (2004) Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA 101(Suppl 2):14572–14579
Bannert N, Kurth R (2006) The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet 7:149–173
Bartolome C, Bello X, Maside X (2009) Widespread evidence for horizontal transfer of transposable elements across Drosophila genomes. Genome Biol 10:R22
Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M (2005) High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol Biol Evol 22:814–817
Berglund J, Pollard KS, Webster MT (2009) Hotspots of biased nucleotide substitutions in human genes. PLoS Biol 7:e26
Best S, Le Tissier PR, Stoye JP (1997) Endogenous retroviruses and the evolution of resistance to retroviral infection. Trends Microbiol 5:313–318
Buzdin A (2007) Human-specific endogenous retroviruses. Sci World J 7:1848–1868
Cann HM, de Toma C, Cazes L, Legrand M-F, Morel V et al (2002) A human genome diversity cell line panel. Science 296:261
Cho K, Lee YK, Greenhalgh DG (2008) Endogenous retroviruses in systemic response to stress signals. Shock 30:105–116
Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10:691–703
de Parseval N, Heidmann T (2005) Human endogenous retroviruses: from infectious elements to human genes. Cytogenet Genome Res 110:318–332
Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D et al (2006) Identification of an infectious progenitor for the multiple-copy HERV-K human endogenous retroelements. Genome Res 16:1548–1556
Dolei A (2006) Endogenous retroviruses and human disease. Expert Rev Clin Immunol 2:149–167
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Feschotte C, Gilbert C (2012) Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13:283–296
Gifford R, Tristem M (2003) The evolution, distribution and diversity of endogenous retroviruses. Virus Genes 26:291–315
Hall TA (1999) BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hughes JF, Coffin JM (2005) Human endogenous retroviral elements as indicators of ectopic recombination events in the primate genome. Genetics 171:1183–1194
Jern P, Coffin JM (2008) Effects of retroviruses on host genome function. Annu Rev Genet 42:709–732
Jha AR, Nixon DF, Rosenberg MG, Martin JN, Deeks SG et al (2011) Human endogenous retrovirus K106 (HERV-K106) was infectious after the emergence of anatomically modern humans. PLoS One 6:e20234
Ji X, Zhao S (2008) DA and Xiao—two giant and composite LTR-retrotransposon-like elements identified in the human genome. Genomics 91:249–258
Kassiotis G (2014) Endogenous retroviruses and the development of cancer. J Immunol 192:1343–1349
Kelley LA, Sternberg MJ (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371
Kent WJ (2002) BLAT—the BLAST-like alignment tool. Genome Res 12:656
Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC et al (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921
Lange T, Deininger MW (2010) Molecular diagnostics in chronic myeloid leukemia. Expert Opin Med Diagn 4:113–124
Lee YN, Bieniasz PD (2007) Reconstitution of an infectious human endogenous retrovirus. PLoS Pathog 3:e10
Lee YK, Chew A, Phan H, Greenhalgh DG, Cho K (2008) Genome-wide expression profiles of endogenous retroviruses in lymphoid tissues and their biological properties. Virology 373:263–273
Lee Y-J, Jeong B-H, Choi E-K, Kim Y-S (2013) Involvement of endogenous retroviruses in prion diseases. Pathogens 2:533–543
Leis J, Baltimore D, Bishop JM, Coffin J, Fleissner E et al (1988) Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol 62:1808–1809
Li X, Slife J, Patel N, Zhao S (2009) Stepwise evolution of two giant composite LTR-retrotransposon-like elements DA and Xiao. BMC Evol Biol 9:128
Lindeskog M, Medstrand P, Cunningham AA, Blomberg J (1998) Coamplification and dispersion of adjacent human endogenous retroviral HERV-H and HERV-E elements; presence of spliced hybrid transcripts in normal leukocytes. Virology 244:219–229
Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS et al (1999) Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73:152–160
López-Sánchez P, Costas JC, Naveira HF (2005) Paleogenomic record of the extinction of human endogenous retrovirus ERV9. J Virol 79:6997–7004
Mills RE, Bennett EA, Iskow RC, Devine SE (2007) Which transposable elements are active in the human genome? Trends Genet 23:183–191
Moyes D, Griffiths DJ, Venables PJ (2007) Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet 23:326–333
Nissen KK, Laska MJ, Hansen B, Terkelsen T, Villesen P et al (2013) Endogenous retroviruses and multiple sclerosis—new pieces to the puzzle. BMC Neurol 13:111
Ogasawara H, Hishikawa T, Sekigawa I, Hashimoto H, Yamamoto N, Maruyama N (2000) Sequence analysis of human endogenous retrovirus clone 4-1 in systemic lupus erythematosus. Autoimmunity 33:15–21
Piotrowski PC, Duriagin S, Jagodzinski PP (2005) Expression of human endogenous retrovirus clone 4-1 may correlate with blood plasma concentration of anti-U1 RNP and anti-Sm nuclear antibodies. Clin Rheumatol 24:620–624
Prusty BK, zur Hausen H, Schmidt R, Kimmel R, de Villiers EM (2008) Transcription of HERV-E and HERV-E-related sequences in malignant and non-malignant human haematopoietic cells. Virology 382:37–45
Repaske R, O’Neill RR, Steele PE, Martin MA (1983) Characterization and partial nucleotide sequence of endogenous type C retrovirus segments in human chromosomal DNA. Proc Natl Acad Sci USA 80:678–682
Repaske R, Steele PE, O’Neill RR, Rabson AB, Martin MA (1985) Nucleotide sequence of a full-length human endogenous retroviral segment. J Virol 54:764–772
Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N (2008) Endogenous retroviruses and cancer. Cell Mol Life Sci 65:3366–3382
Schneider AM, Duffield AS, Symer DE, Burns KH (2009) Roles of retrotransposons in benign and malignant hematologic disease. Cellscience 6:121–145
Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD et al (2005) Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus—specific microarray. J Virol 79:341–352
Shin W, Lee J, Son SY, Ahn K, Kim HS, Han K (2013) Human-specific HERV-K insertion causes genomic variations in the human genome. PLoS One 8:e60605
Smit AF (2008) HERVE_a - ERV1 endogenous retrovirus from Catarrhini. Direct submission to Repbase Update. http://www.girinst.org/repbase/index.html
Steele PE, Rabson AB, Bryan T, Martin MA (1984) Distinctive termini characterize two families of human endogenous retroviral sequences. Science 225:943–947
Takahashi Y, Harashima N, Kajigaya S, Yokoyama H, Cherkasova E et al (2008) Regression of human kidney cancer following allogeneic stem cell transplantation is associated with recognition of an HERV-E antigen by T cells. J Clin Invest 118:1099–1109
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Taruscio D, Floridia G, Zoraqi GK, Mantovani A, Falbo V (2002) Organization and integration sites in the human genome of endogenous retroviral sequences belonging to HERV-E family. Mamm Genome 13:216–222
Verstovsek S (2009) Preclinical and clinical experience with dasatinib in Philadelphia chromosome-negative leukemias and myeloid disorders. Leuk Res 33:617–623
Voisset C, Weiss RA, Griffiths DJ (2008) Human RNA “rumor” viruses: the search for novel human retroviruses in chronic disease. Microbiol Mol Biol Rev 72:157–196
Volkman HE, Stetson DB (2014) The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol 15:415–422
Yang Z (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573
Yi JM, Kim HS (2006) Molecular evolution of the HERV-E family in primates. Arch Virol 151:1107–1116
Yi JM, Kim HS (2007) Molecular phylogenetic analysis of the human endogenous retrovirus E (HERV-E) family in human tissues and human cancers. Genes Genet Syst 82:89–98
Young GR, Eksmond U, Salcedo R, Alexopoulou L, Stoye JP, Kassiotis G (2012) Resurrection of endogenous retroviruses in antibody-deficient mice. Nature 491:774–778
Young GR, Stoye JP, Kassiotis G (2013) Are human endogenous retroviruses pathogenic? An approach to testing the hypothesis. BioEssays 35:794–803
Acknowledgments
We thank C. Phillips and. M. V. Lareu for kindly letting us use their copy of the HGDP-CEPH Human Genome Diversity Panel.
Conflict of interest
The authors declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2014_9789_MOESM2_ESM.pdf
Figure S1. Phylogenetic relationships within the ERVE family based on the analysis of gag, pr, rt and rh gene nucleotide sequences. (PDF 1051 kb)
10709_2014_9789_MOESM3_ESM.pdf
Figure S2. Multiple nucleotide alignment of HCML-ARV with the consensus of subfamily VIIa and the orthologous copies of provirus 022319 in humans and chimpanzees. (PDF 87 kb)
Rights and permissions
About this article
Cite this article
Naveira, H., Bello, X., Abal-Fabeiro, J.L. et al. Evidence for the persistence of an active endogenous retrovirus (ERVE) in humans. Genetica 142, 451–460 (2014). https://doi.org/10.1007/s10709-014-9789-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-014-9789-y