Abstract
Bacterial blight (X. oryzae pv. oryzae) is a serious disease in rice across the world. To better control the disease, it is important to understand its epidemiology and how key aspects of this (e.g. infection efficiency, and spatial spread) change according to environment (e.g. local site conditions and season), management, and in particular, variety resistance. To explore this, we analysed data on the disease progress on resistant and susceptible varieties of rice grown at four sites in the Philippines across five seasons using a combination of mechanistic modelling and statistical analysis. Disease incidence was generally lower in the resistant variety. However, we found no evidence that the primary infection efficiency was lower in resistant varieties, suggesting that differences were largely due to reduced secondary spread. Despite secondary spread being attributed to splash dispersal which is exacerbated by wind and rain, the wetter sites of Pila and Victoria in south Luzon tended to have lower infection rates than the drier sites in central Luzon. Likewise, we found spread in the dry season can be substantial and should therefore not be ignored. In fact, we found site to be a greater determinant of the number of infection attempts suggesting that other environmental and management factors had greater effect on the disease than climate. Primary infection was characterised by spatially-random observations of disease incidence. As the season progressed, we observed an emerging short-range (1.6 m–4 m) spatial structure suggesting secondary spread was predominantly short-range, particularly where the resistant variety was grown.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bacterial leaf blight caused by Xanthomonas oryzae pv. oryzae (Ishiyama, 1922) is one of the most important diseases of rice (Choi et al., 1998; Savary et al., 2000a; Savary et al., 2000b). It is destructive to high-yielding varieties in both temperate and tropical regions, especially in Asia. Early and severe infections of X. oryzae pv. oryzae can result to more than 70% yield loss owing to reductions of physiological (photosynthetic rate, stomatal conductance to CO2, water use efficiency, and leaf transpiration rate) and agronomic components of rice (1000 grain weight and percent filled spikelets; Rao and Kauffman, 1977; Reddy et al., 1979; Reddy and Nayak, 1984; Ou, 1985; Mew et al., 1993; Kumar et al., 2013). Infections at the later booting stage, do not have such a significant effect on yield but result in poor quality grains and a high proportion of broken kernels.
The disease is polycyclic in nature. It survives between seasons on rice stubble and on weeds, considered the primary sources of inoculum. Xanthomonas oryzae is short-lived outside of its host and so does not survive well in the soil and there are contradictory reports of its survival on seed. The pathogen infects wounds and water pores in the plant. It multiplies in intercellular spaces and bacterial ooze is produced on the leaf which can be passed to other plants by direct contact of foliage and through irrigation water (Mundt et al., 1999; Cottyn and Mew, 2004; White and Yang, 2009). Longer-range spread is associated with strong winds and typhoons, which both disperse inoculum and cause wounds that make the plants susceptible in infection (Mundt et al., 1999). Lesions that result from infection still occur on resistant varieties but to a lesser extent, as the pathogen is enveloped by plant polymers making the bacteria unable to colonise the intercellular spaces (Cottyn and Mew, 2004).
Previous reports showed that climatic conditions (temperature at 25 – 30 °C, strong winds, rainfall, and high air humidity) accompanied by inappropriate cultural management (use of susceptible varieties and excessive application of nitrogen) are among the major factors that favour the development of the disease and epidemic spread (Exconde et al., 1973; Horino et al., 1982; Reddy and Nayak, 1984; Ou, 1985; Diekmann and Bogyo, 1992; Savary et al., 1995). The use of resistant varieties remains the most effective and economical control against bacterial leaf blight of rice (Chukwu et al., 2019). This technique commonly starts with breeding either through conventional or molecular approaches. The resistance genes (Xa) widely used in the breeding program for bacterial blight resistance were specific to known races of the pathogen. Of the Xa genes identified from the wild and cultivated rice accessions, race-specific Xa4, xa5, xa13, and Xa21 have been incorporated into modern rice varieties (Singh et al., 2001; Khan et al., 2014; Pradhan et al., 2015). The development and use of modern molecular breeding techniques has fast-tracked gene discovery and has shortened the generation of elite rice lines with novel Xa genes for varietal release from 8-9 years to 3-4 years (Dossa et al., 2015). This means that real-time deployment of resistant varieties with various combinations of resistance genes can be customized through gene rotation or mixture in a single genetic background.
Strategic deployment of bacterial blight-resistant varieties is a sustainable way of achieving long-lasting and stable resistance, in ways that limit the selection of virulent population that may eventually lead to an outbreak. Several studies have shown the effectiveness of Xa genes and deployment methods in slowing down bacterial blight epidemics in the field. Reddy and Nayak (1984) noted a significant reduction of bacterial blight severity in a field planted with a mixture of resistant and susceptible genotypes. Ahmed et al. (1997) found a lower disease severity and incidence on a rice genotype with combinations of Xa genes, particularly Xa4/xa5 pyramid, than the single most effective gene deployed singly. Moreover, Xa7 effectively reduced bacterial blight epidemics in the field even with the presence of Xoo population virulent to the target resistance gene (Ona et al., 1998).
While several studies have demonstrated the effectiveness of Xa genes in slowing down bacterial blight epidemics in the field, the dynamics of bacterial blight development and the processes affecting rates of epidemic development among varieties with resistance genes were not explored. In this study, we set out to compare the effect of resistance genes on the epidemiology of rice bacterial leaf blight and how these might be affected by site and season. To do this, we gathered data on the bacterial blight progress (measured as infection incidence) in field trials from four sites in the Philippines (two in central Luzon and two in southern Luzon) across five consecutive planting seasons. We fitted a model of the disease dynamics to the data and also used statistical analyses to determine the key factors that affect the dynamics of the epidemic. Our objective was to determine to what extent the resistance genes affected the disease incidence, primary and secondary infection efficiency, and the spatial spread of the disease. Additionally, we wanted to determine whether differences between the lines were more affected by site or season.
Materials and Methods
Field experiments
Four monitoring sites for X. oryzae pv. oryzae were established in the Philippines. Two sites were located in the municipalities of Pila (14°13’21”N 121°21’9”E) and Victoria (14°11’6”N 121°20’48”E) in the Laguna province. The other two sites were in the Nueva Ecija province, in the barangays of Malayantoc in the Santo Domingo municipality and Maligaya in the city of Muñoz (located at 15°40’26”N, 120°53’36”E, and 15°40’35”N, 120°52’47”E, respectively; Fig. 1). Xanthomonas oryzae pv. oryzae disease had been previously observed at all four of the experimental sites. Monitoring experiments were conducted for three wet seasons from 2016 to 2018 and two dry seasons in 2017 and 2018. There were six plots at each site. Three were planted with a susceptible line (IR24) and three a pyramided line carrying three Xa resistance genes Xa4+xa5+Xa21 (IRBB57). A total of 400 hills were grown in each plot (40 x 10 hills) with a 20 cm x 20 cm spacing between hills. Resistant and susceptible plots were alternately positioned across the site, with a 20 cm gap between plots, resulting in fields consisting of 40 x 60 hills. Transplanting was done using 21-day old seedlings with three seedlings per hill. In the Nueva Ecija sites a total rate of 72.8 kg N ha-1, in the form of complete fertilizer (14-14-14) was applied in all plots. In the Laguna sites, a total rate of 27.6 kg N ha-1 fertilizer was applied, in the form of urea (46-0-0). The dose at this site was relatively low because a high rate had been applied to these fields in the previous cropping season. All plots were irrigated after transplanting.
The location of experimental sites. The two sites in the Laguna province are located in the municipalities of Pila (14°13’21”N, 121°21’9”E) and Victoria (14°11’6”N, 121°20’48”E). The two sites in the Nueva Ecija province are located in the barangays of Malayantoc in the Santo Domingo municipality (15°40’26”N, 120°53’36”E) and Maligaya in the city of Muñoz (15°40’35”N, 120°52’47”E)
Natural infection of X. oryzae pv. oryzae on rice plants was monitored starting at the late tillering stage (approx. 50-60 days after sowing). After the disease incidence was observed, assessments were done weekly until plants became too senescent to reliably distinguish X. oryzae pv. oryzae lesions. Disease incidence of every hill was recorded and expressed as the number of tillers that were infected per hill (2,400 hills per site).
Analysis with epidemiological models
An epidemiological model simulating the number of infected tillers (I) was fitted to data from both resistant (R) and susceptible (S) varieties simultaneously. The model is given by
where \(\dot{I}(t)\) is the rate of change in the number of infected tillers in each plot; t is the days after transplant (DAT); a represents the rate of infection from primary inoculum; ε represents the decay rate of the primary inoculum’s efficiency due to the combined factors of the rate of decay of the free-living bacteria and the reduction in susceptibility of the rice plant as it matures post-transplant (e.g. due to heightened susceptibility at transplant due to transplant damage); τ is the approximate time when symptoms first show at a monitoring site; and, rS represents the secondary disease transmission rate, i.e. the rate that tillers get infected due to plant-to-plant spread. Process-based epidemiological models, such as this one, have been extensively used to model various plant pathosystems (see Jeger et al. (2018) and references therein). We allowed for parameters a and r to be variety-specific as we expect differences in the infection efficiencies of the pathogen infecting the resistant and susceptible rice varieties. It is biologically plausible that there will be no effect of variety on ε and τ and so to be parsimonious with the model parameters used, we kept these parameters consistent across varieties. The model describes how newly infected tillers may come from one of two sources: primary and secondary inoculum. We model new infections to the resistant and susceptible variety caused due to primary inoculum with the terms aR e−ε(t − τ) and aS e−ε(t − τ), respectively. If ε, rRS and rS are set to zero, then the model reduces to a linear model with slope aR or aS and intercept −aRτ or −aSτ for the resistant or susceptible varieties, respectively. When the decay rate ε > 0, the model begins with a linear increase in infections from primary inoculum but eventually decreases until the primary inoculum is exhausted and no new infections occur. The secondary spread of the disease, which is described by the terms rRS IS(t − τ) and rS IS(t − τ) for the resistant and susceptible varieties, respectively, result in exponential growth of the number of infections. As described above, although the pathogen can cause visible lesions on the leaves of resistant varieties these lesions tend to be small, indicating lower bacterial numbers and reduced ability to become a source of secondary infection (Webb et al., 2010). Therefore, we assume that secondary spread from the resistant variety is negligible, and so the resistant counterpart to rS, rR, is not included in the model. Instead, we include the possibility that the resistant plots may be infected by secondary spread from the susceptible plots that neighbour them at a rate, rRS. These parameters are summarised in Table 1. This model therefore comprises a combination of three types of behaviour (linear, linear with decay and exponential) relating the number of infected tillers over time.
At the end of the observational period there is a drop in the counts of infected tillers in the data (Fig. 2 & 3). This is because diseased tillers are no longer distinguishable from senesced leaves. These points were therefore not considered in the model fitting. The model was fitted to the data using non-linear least squares. Parameters with zero estimates were removed from the model and the reduced model was refitted. In cases where the model fitting did not converge, we further reduced the number of parameters of the model by systematically fitting the model with one of the parameters removed, the reduced model with the best standard error was then selected. We used these statistics in t-tests to determine whether each parameter was significantly different from zero. We analysed these results to make deductions about the epidemiology of X. oryzae pv. oryzae.
The total number of rice tillers per plot infected by Xanthomonas oryzae pv. oryzae. The plots were located at Pila and Victoria in the Laguna province. Two varieties were grown, one resistant to the disease the other susceptible. The points show the observations and the line the model fitted to the data represented by the grey points. Black points show data not used to fit the model
The total number of rice tillers per plot infected by Xanthomonas oryzae pv. oryzae. The plots were located at Malayantoc and Maligaya in the Nueva Ecija province. Two varieties were grown, one resistant to the disease the other susceptible. The points show the observations and the line the model fitted to the data represented by the grey points. Black points show data not used to fit the model
Analysis with statistical models
To reveal the factors that make significant differences in the amount of infection, we fitted Linear Mixed effects Models (LMMs) to the data using the Genstat statistical software package (VSN International, 2019) to total incidence (I(t)) calculated by summing the total number of infected tillers in each plot, resulting in three replicate measurements per treatment. We considered the factors Susceptibility of the crop (with levels susceptible or resistant), Observation number, Season (with levels wet or dry) and Site (with levels Pila, Victoria, Malayantoc and Maligaya) as fixed effects, and Site/Year/Season/Treatment/Observation/Replicate as a random effect. We selected terms using backwards elimination according to the largest p-value given by F-tests. The final predictive model was chosen when all remaining terms gave significant values (P≤0.05) when dropped from the model.
To derive information about spatial structure of the disease and how it changes with crop, season, and location, we fitted variograms to the data on infected tillers across the resistant and susceptible plots for each observation date. The variogram is a tool in spatial statistics that quantifies the spatial dependence in a variable of interest. While it has been widely used in soil science and ecology (Webster and Oliver, 2007; San Martin et al., 2018), it has also been used in plant pathology (Van de Lande and Zadoks, 1999; Bedimo et al., 2007; Nkeng et al., 2017). The variogram is defined as the function that links the expected squared difference of a variable between any two places in a field
where Z(x) and Z(x + h) are random variables at positions x and x + h separated by the vector h for all h. It characterizes quantitatively the spatial dependence in the variable (Webster and Oliver, 2007). The variogram can be estimated by the method of moments
where z(xi) and z(xi + h) are the counts in quadrats centred at xi and xi + h separated by the vector h and m(h) is the number of paired comparisons at that separation. By changing h we obtain an ordered series describing the way the variance changes with the changing separation.
For each set of data, a variogram model was fitted (where possible) using maximum likelihood. For that purpose, we used the likfit directive from the geoR package (Ribeiro Jr et al., 2020) of the R statistical software (R Core Team, 2020). Here we fit a linear mixed model where the fixed effects describe any trend in the data (i.e. the spatial coordinates are the fixed effect variables in the model) and the random spatially correlated component is described with a variogram function. There are few permissible simple models for variograms because the variogram must be such that it cannot return negative variances. In technical terms the model must be conditional negative semi-definite (see Webster and Oliver, 2007). Typically one might test various permissible models to see which fitted the best. Because our aim was to compare model parameters across sites and seasons, after some preliminary testing, we chose to use the exponential model in all cases. This model is defined
where C0 quantifies the spatially uncorrelated variance, known as the nugget, C1 quantifies the spatially correlated part of the sill (or the a priori variance defined C0 + C1 ). The variogram may reach the sill at a finite lag distance known as the range which is approximated by 3d in the exponential model.
First we looked for any evidence of trend across the plots in the data by testing to see if adding a linear fixed effect into the model made a significant improvement (p<0.05). Any evidence of spatial trend would suggest that there were some larger scale effect making one area of the plot more susceptible to disease than another, for example it may show the effect of irrigation channels. We compared the best fitting spatial model (i.e. either the one with or without trend as determined in the previous step) to a model without a spatially correlated random component using the Akaike Information Criterion (AIC). If the data shows little or no spatial component then this implies that the infections are no-more than random, suggesting either limited secondary infection or that the secondary infection is not localised.
We then investigated to see if we could elucidate any general patterns in spatial structure according to crop susceptability, date or site. For this we fitted LMM to the magnitude of spatial structure (C1) and range (estimated as 3d, see Webster and Oliver, 2007) as a function of the factors described above (i.e. Susceptibility of the crop, Observation number, Season and Site as fixed effects and Site/Year/Season/Treatment/Observation as a random effect).
Results
Spatially-resolved plots of the data are available in Fig. S1 – S18 in Online Resource 1. Table 2 reports the maximum number of infected tillers found at a site averaged across each variety’s three replicates. Environmental factors that were believed to affect the disease development of X. oryzae pv. oryzae, such as temperature, relative humidity, and rainfall, were also recorded along with the transplanting date.
Analysis with Epidemiological Models
The model fitted well to each of the sets of data (Fig. 2 & 3). The parameter estimates of the fitted models and the estimate’s significance levels are given in Table 3.
There was no clear effect of site on the significance of aR and aS. While significant values of aR and aS were found in both seasons, they were mostly found in the wet season (Table 3). The only trial where aR and aS were found to be significantly greater than zero during the dry season was at Maligaya in 2017. These estimates were very high, much higher than that found in the significant estimates from the wet seasons, but as these are our only estimates of primary inoculum in the dry season it is not clear if this is typical for the season. Tests were also performed to assess if the aR were significantly less than the aS. In nearly all cases, no significant difference was found between the two. A significant reduction in the resistant variety was only found at Pila during the 2017 wet season. At Maligaya during the 2016 wet season aR was significantly greater than aS, however.
Both rS and rRS were found to be significantly different from zero in many of the cases regardless of site or season. Our estimates of rS were 51% higher on average in the dry season than in the wet season, this is largely due to the very high estimate at Malayantoc during the 2018 dry season. Estimates of rRS were also higher, by 67%, on average. This was again due to a high estimate at Malayantoc during the 2018 dry season. These high estimates are due to the jump in infection that was observed in the final week of sampling.
The time to first symptoms was found to be significant regardless of site or season. It was generally estimated to be between one to four months. The estimates for time to first symptoms were approximately 38 days higher in the dry season than the wet season, and approximately 18 days higher on average at the central Luzon sites than at the south Luzon sites. The estimates of the decay rate parameter, ε, was only significantly greater than zero during the 2017 dry season at Maligaya.
Statistical analysis of incidence levels
Preliminary analysis showed that fitting a model to describe the number of infected tillers resulted in an unacceptable increase in residual variance with increasing number of infected tillers. Therefore, we took natural logarithms of the response variate before fitting our model. The final fitted model describing the number of infected tillers is given by
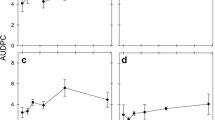
The factor Season and associated interactions were dropped from the model, all other factors were retained. Clearly and not surprisingly the number of infected tillers increased with date of observation and was generally larger in the susceptible variety compare to the resistant at the same site-season (Fig. 4). Pila and Victoria tended to have lower infection rates, notably in resistant varieties. The largest predictions of infection were related to Malayantoc and Maligaya.
Analysis with spatial structure
We assume evidence of spatial trend in the disease incidence if the fixed effects in the LMM (which describe linear trend) explains a significant part of the variation (p < 0.05). There was little significant evidence of spatial trend in the disease incidence across observations, except for at Malayantoc where in all but one season at least a third of observations showed evidence of spatial trend (Table 4). Fig. 5 shows the variogram fits for the susceptible plots at Malayantoc in the wet season of 2016. Here, four of the eight observations had evidence of significant spatial trend (Table 4). This is also visually evident from variogram models (weeks 4, 6 – 8). If trend is present in the data (and unaccounted for) then typically the variogram does not rise to an asymptote but continues to increase with increasing lag distance. Visual inspection of the empirical variograms (shown by the points; see Fig. 5 and additional examples in Fig. S19 – S54 in Online Resource 2) shows that this is often related to a secondary process with lag distance just over 2 m (the second rise in the variogram), which is the distance between plots of a similar type. This might relate to variation in environmental conditions affecting the diseases as well as secondary dispersal.
Susceptible variety at Malayantoc in wet season 2016. Each pane shows the results from eight observations taken across the season. The points show the empirical variogram calculated using the method of moments, the grey line is the variogram model fitted without a trend effect accounted for and the black line is the variogram model with the trend accounted for. We note that neither line is a fit to the points shown, all three relate to different methods for fitting the variogram to the data set
We compared the best fitting spatial model (i.e. either the one with or without trend as determined in the previous step) to a model without a spatially correlated random component using the Akaike Information Criterion (AIC). We found that there was significant spatial structure in the data for almost all observations made at Malayantoc and Maligaya, whereas for data from Pila and Victoria there were more occasions where the random model fitted the data better than the model with spatial structure (Table 5) this was particularly true in the resistant varieties and suggests there was no evidence of secondary infection. We generally found little to no spatial structure in the first observations of the plots across sites and seasons. In many cases the “nugget” model (indicated by near constant variance with increasing lag) gave the best fit (see Fig. 6 for an example). This suggests that primary inoculum infects the crop in a random fashion.
Variograms of the incidence of X. oryzae pv. oryzae in (a–f) resistant and (g–l) susceptible plots observed on six occasions at the Victoria site in the wet season of 2016. The points show the empirical variogram, the grey line is the variogram model fitted without a trend effect and the black line is the variogram model with the trend accounted for. The variograms for the resistant variety show a flat structure that persists over time suggesting little secondary infection. The variograms for the susceptible variety show an increase in spatial structure over time indicating local spatial spread
Preliminary analysis showed that fitting linear models to describe the magnitude of the spatial component of the variograms across the sites (C1) and the distance parameter (d) resulted in an unacceptable increase in residual variance with increasing value of the response variate. Therefore, in both cases we took natural logarithms of the response variate before fitting our model. The final fitted model for the model of the spatial component is given by
All terms were retained in the full model with some higher order interactions dropped. The model showed that the spatial structure quantified by the C1 parameter tended to be smaller in the resistant plots compared with the susceptible plots (see Fig. S55 in Online Resource 2) across sites. Season had no comparable consistent affect across sites. There was an increase in C1 with time at all sites. The increase in structure is evidence of secondary spread, although we note that the random component (the nugget) was often relatively large.
Only the site parameter was retained in the model to explain the variation in range, no other factors were significant in the model. Malayantoc showed the largest predicted range parameter suggesting that the correlated structure extended up to 6m (Table 6). This is likely to reflect the larger scale differences across the plots identified by the variogram models with trend included. The range of the correlated structure was predicted to be smallest at the Laguna sites with a predicted mean value of 1.64 m for Victoria. This is likely to reflect the short-scale transmission of the disease.
Discussion
Here we have described, with epidemiological and statistical models, how the resistance genes affected the bacterial blight incidence, primary and secondary infection efficiency, and the spatial spread of the disease. We have further clarified how any differences in disease dynamics between the resistant and susceptible varieties were affected by site and season. This experiment is unique in that every hill was assessed across multiple time points allowing for an in-depth analysis both temporally and spatially. To be able to assess every hill, we decided to sample the number of infected tillers, rather than assess infected leaf area. Sampling at this level allowed us to sample a site within a day while providing a sufficient level of data for modelling purposes.
Across sites and seasons the resistant varieties showed less disease incidence than the susceptible varieties (Fig. 2 & 3). At some sites and seasons the incidence levels on resistant varieties were substantial. However, lesions on these plants were small (less than 5cm) indicating that the resistant variety is still effective against the local pathogen population and that secondary spread from these plants were likely to be limited. Indeed, the spatial analysis showed that in most cases there was no indication of spatial structure in the resistant plots supporting the observation that secondary spread was unlikely to have occurred between resistant plants. We note however, that due to the close proximity of the susceptible plots, infection attempts on the resistant variety from secondary inoculum from the susceptible varieties were likely, as reflected in our model formulation and supported by the significant rRS parameters values obtained in many of the site-seasons.
We observed three general patterns in the increase of infected tillers over time: (i) linear increase (e.g. Maligaya wet season 2018); (ii) asymptotic increase (e.g. Maligaya dry season 2017); and, (iii) exponential increase (e.g. Malayantoc wet season 2016). This corresponds to the three types of dynamics the model can describe and indicates the dominant processes responsible for the observed data. Linear increase depends only on the rate of infection from the primary inoculum. Asymptotic increase occurs when the rate of infection decreases over time, e.g. due to inoculum levels decreasing or reduction in susceptibility of the plant. In our model this behaviour is governed by a rate of decay parameter acting upon the primary inoculum. Exponential increase of infection occurs when the disease is spread via inoculum produced from neighbouring plants, i.e. secondary spread.
The dynamics of the model are such that for infection to occur on the susceptible variety there must be primary infection on the susceptible variety whereas the resistant variety can be infected either by primary inoculum or from secondary spread from the susceptible variety. This means that the parameter aS must be greater than zero, whereas the parameter aR may be greater than or equal to zero. Despite this, there were many cases where aS was not significantly greater than zero. In several cases, significant secondary infection made it difficult to detect significant primary inoculum, which may explain why so few cases were found to be significant. For example, only in Maligaya during the 2016 and 2017 wet seasons were both aS and rS significant, and only in the latter were both aR and rRS significant. Our estimates of aR were generally not found to be significantly different from aS, suggesting that, the primary inoculum had a similar effect on the number of infected tillers of the susceptible variety as the resistant. This means that resistance doesn’t reduce the number of lesions caused by primary inoculum significantly. Given that we still see less infection in the resistant variety, suggests that resistance leads to a lack of secondary spread, supporting the results from the statistical analysis.
Weather, particularly rainfall and windspeed, is believed to be a main driver of secondary spread (Naqvi, 2019 and references therein), however this was not supported by our results. At some sites we found evidence of significant secondary spread even in very dry conditions (e.g. at Malayantoc and Maligaya during the 2018 dry season) and no evidence for secondary spread at other sites in wetter conditions (e.g. at Malayantoc and Maligaya during the 2018 wet season), see Table 5.
Our analysis of the data using LMMs showed that site had a more significant effect on disease incidence than season (addressing our secondary objective). We found the northern sites in Nueva Ecija (Malayantoc and Maligaya) generally showed greater levels of disease incidence than those in the south, yet these sites had less rainfall and lower windspeeds than those in the south (Table 2). All sites had average temperatures in the range considered optimal for the disease (25-30°C; Naqvi, 2019). Crop management is also reported to significantly influence disease, and it is likely that this is a key driver in the differences in incidence that we observed in our experiments. In particular nitrogen fertilizer is reported to positively correlate with X. oryzae pv. oryzae and we found that the disease incidence was greatest at Malayantoc and Maligaya where a total of 72.8 kg/ha was applied in each season compared with Pila and Victoria where only 27.6 kg/ha was applied. Although the Nueva Ecija sites have higher rate of nitrogen application than in the Laguna sites, the 72.8 kg/ha in the Nueva Ecija sites is still lower than the recommended rate that farmers are told to use to avoid high incidence of bacterial blight. That is to say, the rate of nitrogen application in the Nueva Ecija sites is not considered high in the context of that area and may not necessarily explain the greater incidence observed among the Nueva Ecija sites as compared with those in the Laguna sites. Further, the low fertiliser rate applied in the south was in response to a high application in the previous cropping season (as explained above). As nitrogen fertilizer rate was somewhat confounded by location, we did not include this as a factor in our statistical model.
The model-based estimates suggested it took an average of two months after transplantation for symptoms of X. oryzae pv. oryzae to appear. The infection took the least amount of time to appear in the 2017 wet-season, where symptoms were predicted as early as one month. Two site-seasons had predicted infection times of over three months. A study by Mundt et al. (1999) investigating plant-to-plant spread from a clip-inoculated plant suggested that symptoms should develop in neighbouring plants within 30-35 days after inoculation of the inoculated plant. We would expect that plants naturally infected by primary inoculum would take longer for symptoms to develop on the leaf. Our average estimate of two months for symptoms to develop from transplant may therefore not be unreasonable.
We found little evidence that the source of primary inoculum decays over time or that plants become less susceptible, as the effect was only found to be significant at Maligaya during the 2017 dry season. This may be due to other more dominant processes, e.g. secondary spread masking this effect or it may suggest that either the bacteria can survive on weeds and the rice stubble of previous crops for long periods of time or that there is a significant amount of primary inoculum replenishing the decaying pathogen population.
Disease infection in crops may appear spatially random or may have a more aggregated pattern. Generally, spatial aggregation suggests that the pathogen spreads locally from plant to plant, whereas a random pattern suggests other larger-scale processes dominate (Nayak and Reddy, 1987). Mundt et al. (1999), report that although epidemics of X. oryzae pv. oryzae can begin from small foci of initial infection, the initial disease pattern can appear to be random. Our spatial analysis suggests that the primary inoculum infects the plots in a random fashion (as indicated by the variograms fitted to the data from the first observations made at each site-season). Thereafter, in most site-seasons evidence of spatial structure emerged. This suggests local secondary spread. Our observations support the findings of Nayak and Reddy (1987) who observed random patterns of infection at low incidence with aggregation occurring at higher levels of disease as secondary spread takes hold. The variogram range parameter quantifies the extent of local spatial dependence in disease incidence. Malayantoc showed the largest predicted range parameter suggesting that the correlated structure extended up to 6m (Table 6). Closer inspection of the variogram however suggests that this larger scale process is likely to reflect the differences between plots at that site (Fig. 5). The shorter-range process at this site suggests a correlated structure with less than 2m-range. The range of the correlated structure was predicted to be smallest at the Laguna sites with a mean value of 1.64 m for Victoria and 3.18 m for Pila. These relatively short-range parameters are not surprising. Splash dispersal of X. oryzae pv. oryzae is reported to be quite a localised phenomenon with droplets estimated to generally disperse between 0.2 and 1 m (Mundt et al., 1999). Longer distance dispersal tends to be attributed to smaller splash droplets being dispersed by the wind (Ou, 1985; Mundt et al., 1999).
In this article, we show how epidemiological and statistical models can be used in tandem with large data sets on disease to reveal new insights into the mechanisms of the epidemic. Our analysis showed primary infection in resistant varieties was not significantly different from susceptible varieties and that there was little evidence to suggest that the source of primary infection decayed over the season. However, the secondary spread of disease infection was not evident among the resistant variety. Primary infection appeared spatially random. As the season progressed, at many sites we observed an emerging short-range (1.6 m – 4 m) spatial structure in the disease incidence suggesting secondary spread was predominantly short-range. The spatially structured variation tended to be smaller in the plots where the resistant variety was grown. All these findings highlighted the important role of resistance genes in rice bacterial blight management by effectively slowing down disease epidemics in the field. While bacterial blight incidence is commonly high during the wet cropping season, our results have shown that the disease can also effectively established during the dry cropping season. For the sites considered, we found climate variables were not a good predictor of infection suggesting that other environmental and management factors (such as fertilizer application) had greater effect.
References
Ahmed, H. U., Finckh, M. R., Alfonso, R. F., & Mundt, C. C. (1997). Epidemiological effect of gene deployment strategies on bacterial blight of rice. Phytopathology, 87(1), 66–70. https://doi.org/10.1094/phyto.1997.87.1.66
Bedimo, J. A. M., Bieysse, D., Cilas, C., & Notteghem, J. L. (2007). Spatio-temporal dynamics of arabica coffee berry disease caused by Colletotrichum kahawae on a plot scale. Plant Disease, 91(10), 1229–1236. https://doi.org/10.1094/pdis-91-10-1229
Choi, S. H., Cruz, C. M. V., & Leach, J. E. (1998). Distribution of Xanthomonas oryzae pv. oryzae DNA modification systems in Asia. Applied and Environmental Microbiology, 64(5), 1663–1668. https://doi.org/10.1128/AEM.64.5.1663-1668.1998
Chukwu, S. C., Rafii, M. Y., Ramlee, S. I., Ismail, S. I., Hasan, M. M., Oladosu, Y. A., Magaji, U. G., Akos, I., & Olalekan, K. K. (2019). Bacterial leaf blight resistance in rice: a review of conventional breeding to molecular approach. Molecular Biology Reports, 46(1), 1519–1532. https://doi.org/10.1007/s11033-019-04584-2
Cottyn, B. & Mew, T. (2004). Bacterial Blight of Rice. In: R. M. Goodman (Ed.), Encyclopedia of Plant and Crop Science. (1st ed., pp. 79-83). CRC Press.
Diekmann, M., & Bogyo, T. P. (1992). Distribution of bacterial leaf-blight of rice Xanthomonas-campestris pv oryzae (Ishiyama) dye depending on climatological factors. Zeitschrift Fur Pflanzenkrankheiten Und Pflanzenschutz-Journal of Plant Diseases and Protection, 99(2), 127–136.
Dossa, G. S., Sparks, A., Cruz, C. V., & Oliva, R. (2015). Decision tools for bacterial blight resistance gene deployment in rice-based agricultural ecosystems. Frontiers in Plant Science, 6. https://doi.org/10.3389/fpls.2015.00305
Exconde, O. R., Opina, O. S., & Phanomsawaran, A. (1973). Yield losses due to bacterial leaf blight of rice. Philippine Agriculturist, 57, 128–140.
Horino, O., Mew, T. W., & Yamada, T. (1982). The effect of temperature on the development of bacterial leaf blight on rice. Japanese Journal of Phytopathology, 48(1), 72–75. https://doi.org/10.3186/jjphytopath.48.72
Ishiyama, S. (1922). Studies of bacterial leaf blight of rice. Report of the Imperial Agricultural Station, Nishigahara (Konosu), 45, 233–261.
Jeger, M. J., Madden, L. V., & van den Bosch, F. (2018). Plant Virus Epidemiology: Applications and Prospects for Mathematical Modeling and Analysis to Improve Understanding and Disease Control. Plant Dis., 102(5), 837–854. https://doi.org/10.1094/pdis-04-17-0612-fe
Khan, M. A., Naeem, M., & Iqbal, M. (2014). Breeding approaches for bacterial leaf blight resistance in rice (Oryza sativa L.), current status and future directions. Eur. J. Plant Pathol., 139(1), 27–37. https://doi.org/10.1007/s10658-014-0377-x
Kumar, A., Guha, A., Bimolata, W., Reddy, A., Laha, G. S., Sundaram, R., Pandey, M., & Ghazi, I. (2013). Leaf gas exchange physiology in rice genotypes infected with bacterial blight: An attempt to link photosynthesis with disease severity and rice yield. Australian Journal of Crop Science, 7, 32–39.
Mew, T. W., Alvarez, A. M., Leach, J. E., & Swings, J. (1993). Focus on bacterial-blight of rice. Plant Disease, 77(1), 5–12. https://doi.org/10.1094/pd-77-0005
Mundt, C. C., Ahmed, H. U., Finckh, M. R., Nieva, L. P., & Alfonso, R. F. (1999). Primary disease gradients of bacterial blight of rice. Phytopathology, 89(1), 64–67. https://doi.org/10.1094/phyto.1999.89.1.64
Naqvi, S. A. H. (2019). Bacterial leaf blight of rice: An overview of epidemiology and management with special reference to Indian sub-continent. Pakistan Journal of Agricultural Research, 32(2), 359–380.
Nayak, P., & Reddy, P. R. (1987). The pattern of bacterial leaf-blight disease development and spread in rice. Journal of Phytopathology-Phytopathologische Zeitschrift, 119(3), 255–261. https://doi.org/10.1111/j.1439-0434.1987.tb04396.x
Nkeng, M. N., Mousseni, I. B. E., Nomo, L. B., Sache, I., & Cilas, C. (2017). Spatio-temporal dynamics on a plot scale of cocoa black pod rot caused by Phytophthora megakarya in Cameroon. Eur. J. Plant Pathol., 147(3), 579–590. https://doi.org/10.1007/s10658-016-1027-2
Ona, I., Cruz, C. M. V., Nelson, R. J., Leach, J. E., & Mew, T. W. (1998). Epidemic development of bacterial blight on rice carrying resistance genes Xa-4, Xa-7, and Xa-10. Plant Disease, 82(12), 1337–1340. https://doi.org/10.1094/pdis.1998.82.12.1337
Ou, S. H. (1985). Rice Diseases (2nd ed.). Commonwealth Mycological Institute.
Pradhan, S. K., Nayak, D. K., Mohanty, S., Behera, L., Barik, S. R., Pandit, E., Lenka, S., & Anandan, A. (2015). Pyramiding of three bacterial blight resistance genes for broad-spectrum resistance in deepwater rice variety. Jalmagna. Rice, 8, 19. https://doi.org/10.1186/s12284-015-0051-8
R Core Team. (2020). R: A language and environment for statistical computing. R Foundation for Statistical Computing https://www.R-project.org/
Rao, P. S., & Kauffman, H. E. (1977). Potential yield losses in dwarf rice varieties due to bacterial-blight in India. Phytopathologische Zeitschrift-Journal of Phytopathology, 90(3), 281–284. https://doi.org/10.1111/j.1439-0434.1977.tb03247.x
Reddy, A. P. K., Mackenzie, D. R., Rouse, D. I., & Rao, A. V. (1979). Relationship of bacterial leaf-blight severity to grain-yield of rice. Phytopathology, 69(9), 967–969.
Reddy, P. R., & Nayak, P. (1984). Progress of bacterial leaf-blight disease in mixed population of resistant and susceptible rice cultivars. Acta Phytopathologica Academiae Scientiarum Hungaricae, 19(3-4), 285–290.
Ribeiro Jr., P. J., Diggle, P. J., Schlather, M., Bivand, R., & Ripley, B. (2020). geoR: Analysis of Geostatistical Data. R package version, 1, 8–1 https://CRAN.R-project.org/package=geoR
San Martin, C., Milne, A. E., Webster, R., Storkey, J., Andujar, D., Fernandez-Quintanilla, C., & Dorado, J. (2018). Spatial Analysis of Digital Imagery of Weeds in a Maize Crop. ISPRS International Journal of Geo-Information, 7(2), 61. https://doi.org/10.3390/ijgi7020061
Savary, S., Castilla, N. P., Elazegui, F. A., McLaren, C. G., Ynalvez, M. A., & Teng, P. S. (1995). Direct and indirect effects of nitrogen supply and disease source structure on rice sheath blight spread. Phytopathology, 85(9), 959–965. https://doi.org/10.1094/Phyto-85-959
Savary, S., Willocquet, L., Elazegui, F. A., Castilla, N. P., & Teng, P. S. (2000a). Rice pest constraints in tropical Asia: Quantification of yield losses due to rice pests in a range of production situations. Plant Disease, 84(3), 357–369. https://doi.org/10.1094/pdis.2000.84.3.357
Savary, S., Willocquet, L., Elazegui, F. A., Teng, P. S., Van Du, P., Zhu, D. F., Tang, Q. Y., Huang, S. W., Lin, X. Q., Singh, H. M., & Srivastava, R. K. (2000b). Rice pest constraints in tropical Asia: Characterization of injury profiles in relation to production situations. Plant Disease, 84(3), 341–356. https://doi.org/10.1094/pdis.2000.84.3.341
Sharp, R. T. (2021). NFRR_Philippines (Version 1.0.0). Zenodo. https://doi.org/10.5281/zenodo.5786514
Singh, S., Sidhu, J. S., Huang, N., Vikal, Y., Li, Z., Brar, D. S., Dhaliwal, H. S., & Khush, G. S. (2001). Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theoretical and Applied Genetics, 102(6-7), 1011–1015. https://doi.org/10.1007/s001220000495
Van de Lande, H. L., & Zadoks, J. C. (1999). Spatial patterns of spear rot in oil palm plantations in Surinam. Plant Pathology, 48(2), 189–201.
VSN International (2019). Genstat for Windows 20th Edition. VSN International, Hemel Hempstead, UK. Genstat.co.uk.
Webb, K. M., Garcia, E., Cruz, C. M. V., & Leach, J. E. (2010). Influence of rice development on the function of bacterial blight resistance genes. Eur. J. Plant Pathol., 128(3), 399–407. https://doi.org/10.1007/s10658-010-9668-z
Webster, R. & Oliver, M. A. (2007). Geostatistics for Environmental Scientists (2nd ed.). Wiley.
White, F. F., & Yang, B. (2009). Host and pathogen factors controlling the Rice-Xanthomonas oryzae interaction. Plant Physiology, 150(4), 1677–1686. https://doi.org/10.1104/pp.109.139360
Availability of data and material
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Code Availability
The code is available from Zenodo (Sharp, 2021) at https://doi.org/10.5281/zenodo.5786514.
Funding
This work was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) Newton Fund [grant number BB/N01362X/1] and supported by funds from the Department of Agriculture - Philippine Rice Research Institute. AEM was supported by the Institute Strategic Programme Grants ‘Smart Crop Protection’ [grant number BBS/OS/CP/000001], funded by BBSRC. Rothamsted Research receives support from BBSRC.
Author information
Authors and Affiliations
Contributions
Conceptualization: Jennifer T. Niones, Ryan T. Sharp, Ricardo Oliva; Methodology: Jennifer T. Niones, Ryan T. Sharp, Eula Gems M. Oreiro, Alice E. Milne; Software: Ryan T. Sharp; Validation: Jennifer T. Niones, Ryan T. Sharp, Dindo King M. Donayre, Eula Gems M. Oreiro, Alice E. Milne; Formal analysis: Ryan T. Sharp, Alice E. Milne; Investigation: Jennifer T. Niones, Dindo King M. Donayre, Eula Gems M. Oreiro; Resources: Jennifer T. Niones, Dindo King M. Donayre, Eula Gems M. Oreiro, Ricardo Oliva; Data Curation: Ryan T. Sharp; Writing – Original Draft: Ryan T. Sharp, Dindo King M. Donayre, Alice E. Milne; Writing – Review & Editing: Jennifer T. Niones, Ryan T. Sharp, Dindo King M. Donayre, Eula Gems M. Oreiro, Alice E. Milne, Ricardo Oliva; Visualization: Ryan T. Sharp, Alice E. Milne; Supervision: Jennifer T. Niones, Ricardo Oliva, Alice E. Milne; Project administration: Jennifer T. Niones, Alice E. Milne, Ricardo Oliva; Funding acquisition: Jennifer T. Niones, Ryan T. Sharp, Ricardo Oliva.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Niones, J.T., Sharp, R.T., Donayre, D.K.M. et al. Dynamics of bacterial blight disease in resistant and susceptible rice varieties. Eur J Plant Pathol 163, 1–17 (2022). https://doi.org/10.1007/s10658-021-02452-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-021-02452-z