Abstract
Background
DNA methylation mainly affects tumor suppressor genes in the development of hepatocellular carcinoma (HCC). However, sera methylation of specific genes in hepatitis B virus (HBV)-related HCC remains unknown.
Aims
The purpose of this study was to identify methylation frequencies of sera E-cadherin (CDH1), DNA methyltransferase 3b (DNMT3b) and estrogen receptor 1 (ESR1) promoter in HBV-related HCC and analyze the associated clinical significance.
Methods
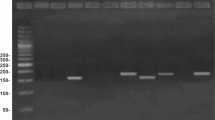
Methylation-specific PCR was used to determine the frequencies of DNA methylation for CDH1, DNMT3b and ESR1 genes in sera from 183 patients with HCC, 47 liver cirrhosis (LC), 126 chronic hepatitis B (CHB), and 50 normal controls (NCs).
Results
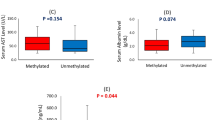
Significantly higher frequencies of methylation of CDH1, DNMT3b and ESR1 were found in HBV-related HCC compared with LC, CHB and NCs. Nodule numbers, tumor size and the presence of liver cirrhosis were significantly associated with gene methylation status in HBV-related HCC. Moreover, HBV may have a strong and enhanced effect on the concurrent methylation of CDH1, DNMT3b and ESR1 in HBV-related HCC. More importantly, combined methylation as a biomarker displayed significantly higher diagnostic value than AFP to discriminate HCC from CHB and LC.
Conclusions
Aberrant sera DNA methylation of CDH1, DNMT3b and ESR1 gene promoters could be a biomarker in the early diagnosis of HBV-related HCC.




Similar content being viewed by others
References
Yuan SX, Yang F, Yang Y, et al. Long noncoding RNA associated with microvascular invasion in hepatocellular carcinoma promotes angiogenesis and serves as a predictor for hepatocellular carcinoma patients’ poor recurrence-free survival after hepatectomy. Hepatology. 2012;56:2231–2241.
Marrero JA, Feng Z, Wang Y, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology. 2009;137:110–118.
Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022.
Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492.
Kim JS, Han J, Shim YM, Park J, Kim DH. Aberrant methylation of H-cadherin (CDH13) promoter is associated with tumor progression in primary nonsmall cell lung carcinoma. Cancer. 2005;104:1825–1833.
Perret C. Methylation profile as a new tool for classification of hepatocellular carcinoma. J Hepatol. 2014;54:602–603.
Venturelli S, Berger A, Weiland T, et al. Differential induction of apoptosis and senescence by the DNA methyltransferase inhibitors 5-azacytidine and 5-aza-2′-deoxycytidine in solid tumor cells. Mol Cancer Ther. 2013;12:2226–2236.
Xu B, Di J, Wang Z, et al. Quantitative analysis of RASSF1A promoter methylation in hepatocellular carcinoma and its prognostic implications. Biochem Biophys Res Commun. 2013;438:324–328.
Kim JG, Takeshima H, Niwa T, et al. Comprehensive DNA methylation and extensive mutation analyses reveal an association between the CpG island methylator phenotype and oncogenic mutations in gastric cancers. Cancer Lett. 2013;330:33–40.
Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007;110:2785–2792.
Zmetakova I, Danihel L, Smolkova B, et al. Evaluation of protein expression and DNA methylation profiles detected by pyrosequencing in invasive breast cancer. Neoplasma. 2013;60:635–646.
Huidobro C, Urdinguio RG, Rodriguez RM, et al. A DNA methylation signature associated with aberrant promoter DNA hypermethylation of DNMT3B in human colorectal cancer. Eur J Cancer. 2012;48:2270–2281.
Hishida M, Nomoto S, Inokawa Y, et al. Estrogen receptor 1 gene as a tumor suppressor gene in hepatocellular carcinoma detected by triple-combination array analysis. Int J Ooncol. 2013;43:88–94.
Brechot C. Pathogenesis of hepatitis B virus-related hepatocellular carcinoma: old and new paradigms. Gastroenterology. 2004;127:s56–s61.
Quetier I, Brezillon N, Duriez M, et al. Hepatitis B virus HBx protein impairs liver regeneration through enhanced expression of IL-6 in transgenic mice. J Hepatol. 2013;59:285–291.
Lee JO, Kwun HJ, Jung JK, Choi KH, Min DS, Jang KL. Hepatitis B virus X protein represses E-cadherin expression via activation of DNA methyltransferase 1. Oncogene. 2005;24:6617–6625.
Zheng DL, Zhang L, Cheng N, et al. Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J Hepatol. 2009;50:377–387.
Bruix J, Sherman M. Management of hepatocellular carcinoma. Hepatology. 2005;42:1208–1236.
Yasuda H, Soejima K, Nakayama S, et al. Bronchoscopic microsampling is a useful complementary diagnostic tool for detecting lung cancer. Lung Cancer. 2011;72:32–38.
Qian ZR, Sano T, Yoshimoto K, et al. Tumor-specific downregulation and methylation of the CDH13 (H-cadherin) and CDH1 (E-cadherin) genes correlate with aggressiveness of human pituitary adenomas. Mod Pathol. 2007;20:1269–1277.
Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. ChemBioChem. 2011;12:206–222.
Zhu J, Du S, Zhang J, et al. Polymorphism of DNA methyltransferase 3B-149C/T and cancer risk: a meta-analysis. Med Oncol. 2015;32:399.
Jung JK, Arora P, Pagano JS, Jang KL. Expression of DNA methyltransferase 1 is activated by hepatitis B virus X protein via a regulatory circuit involving the p16INK4a-cyclin D1-CDK 4/6-pRb-E2F1 pathway. Cancer Res. 2007;67:5771–5778.
van Roy F, Berx G. The cell–cell adhesion molecule E-cadherin. Cell Mol Life Sci. 2008;65:3756–3788.
Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1:a003129.
Rodriguez FJ, Lewis-Tuffin LJ, Anastasiadis PZ. E-cadherin’s dark side: possible role in tumor progression. Biochim Biophys Acta. 2012;826:23–31.
Shimizu IKN, Tamaki K, Shono M, Huang HW, He JH, Yao DF. Female hepatology: favorable role of estrogen in chronic liver disease with hepatitis B virus infection. World J Gastroenterol. 2007;32:4295–4305.
Kamalakaran S, Varadan V, Giercksky Russnes HE, et al. DNA methylation patterns in luminal breast cancers differ from non-luminal subtypes and can identify relapse risk independent of other clinical variables. Mol Oncol. 2011;5:77–92.
Wei X, Xiang T, Ren G, et al. miR-101 is down-regulated by the hepatitis B virus X protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013;25:439–446.
Lambert MP, Paliwal A, Vaissière T, et al. Aberrant DNA methylation distinguishes hepatocellular carcinoma associated with HBV and HCV infection and alcohol intake. J Hepatol. 2011;54:705–715.
Ministry of Health PsRoC. The management of primary liver cancer: update 2011. Chin Clin Oncol. 2011;16:929–946.
Acknowledgments
This work was supported by grants from the Science and Technology Development Plan of Shandong Province (2014GSF118068), the National Natural Science Foundation of China (81171579, 81201287, 81371832), and the Key Project of Chinese Ministry of Science and Technology (2012ZX10002007, 2013ZX10002001).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Dou, CY., Fan, YC., Cao, CJ. et al. Sera DNA Methylation of CDH1, DNMT3b and ESR1 Promoters as Biomarker for the Early Diagnosis of Hepatitis B Virus-Related Hepatocellular Carcinoma. Dig Dis Sci 61, 1130–1138 (2016). https://doi.org/10.1007/s10620-015-3975-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-015-3975-3