Abstract
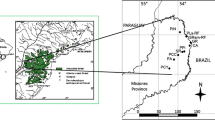
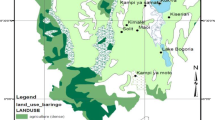
Human activities such as fragmentation and selective logging of forests can threaten population viability by modification of ecological and genetic processes. Using six microsatellite markers, we examined the effects of forest fragmentation and local disturbance on the genetic diversity and structure of adult trees (N = 110) and seedlings (N = 110) of Prunus africana in Kakamega Forest, western Kenya. Taking samples of adults and seedlings allowed for study of changes in genetic diversity and structure between generations. Thereby, adults reflect the pattern before and seedlings after intensive human impact. Overall, we found 105 different alleles in the six loci examined, 97 in adults and 88 in seedlings. Allelic richness and heterozygosity were significantly lower in seedlings than in adults. Inbreeding increased from adult tree to seedling populations. Genetic differentiation of adult trees was low (overall F ST = 0.032), reflecting large population sizes and extensive gene flow in the past. Genetic differentiation of seedlings was slightly higher (overall F ST = 0.044) with all of the 28 pairwise F ST-values being significantly different from zero. These results suggest that human disturbance in Kakamega Forest has significantly reduced allelic richness and heterozygosity, increased inbreeding and slightly reduced gene flow in P. africana in the past 80–100 years.


Similar content being viewed by others
References
Aldrich PR, Hamrick JL (1998) Reproductive dominance of pasture trees in a fragmented tropical forest mosaic. Science 281:103–105
Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G (1998) Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Mol Ecol 7:933–944
Alvarez-Buylla ER, Chaos A, Pinero D, Garay AA (1996) Demographic genetics of a pioneer tropical tree species: patch dynamics, seed dispersal and seed banks. Evolution 50:1155–1166
Blackett HL (1994) Forest inventory report no. 3: Kakamega Kenya indigenous forest conservation programme. Nairobi, Kenya
Bleher B, Uster D, Bergsdorf T (2006) Assessment of threat status and management effectiveness in Kakamega Forest, Kenya. Biodivers Conserv 15:1159–1177
Brooks TM, Pimm SL, Oyugi JO (1999) Time lag between deforestation and bird extinction in tropical forest fragments. Conserv Biol 13:1140–1150
Cespedes M, Gutierrez MV, Holbrook NM, Rocha OJ (2003) Restoration of genetic diversity in the dry forest tree Swietenia macrophylla (Meliaceae) after pasture abandonment in Costa Rica. Mol Ecol 12:3201–3212
Chapin FS, Zavaleta ES, Eviner VT et al (2000) Consequences of changing biodiversity. Nature 405:234–242
Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R (1999) AC/GT and AG/CT microsatellite repeats in peach Prunus persica (L) Batsch: isolation, characterisation and cross-species amplification in Prunus. Theor App Genet 99:65–72
Cloutier D, Kanashiro M, Ciampi AY, Schoen DJ (2007) Impact of selective logging on inbreeding and gene dispersal in an Amazonian tree population of Carapa guianensis Aubl. Mol Ecol 16:797–809
Cunningham AB, Mbenkum FT (1993) Sustainability of harvesting Prunus africana bark in Cameroon. People and Plants Working Paper, 2. UNESCO, Paris
Dawson IK, Powell W (1999) Genetic variation in the Afromontane tree Prunus africana, an endangered medicinal species. Mol Ecol 8:151–156
Dayanandan S, Dole J, Bawa K, Kesseli R (1999) Population structure delineated with microsatellite markers in fragmented populations of a tropical tree, Carapa guianensis (Meliaceae). Mol Ecol 8:1585–1592
Doligez A, Joly HI (1997) Genetic diversity and spatial structure within a natural stand of a tropical forest tree species, Carapa procera (Meliaceae), in French Guiana. Heredity 79:72–82
Excoffier L, Laval G, Schneider S (2005) arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol Bioinfor Online 1:47–50
Farwig N, Böhning-Gaese K, Bleher B (2006) Enhanced seed dispersal of Prunus africana in fragmented and disturbed forests? Oecologia 147:238–252
Fashing PJ (2004) Mortality trends in the African cherry (Prunus africana) and the implications for colobus monkeys (Colobus guereza) in Kakamega Forest, Kenya. Biol Conserv 120:449–459
Fernandez-M JF, Sork VL (2007) Genetic variation in fragmented forests stands of the Andean oak Quercus humboldtii Bonpl. (Fagaceae). Biotropica 39:72–78
Godoy JA, Jordano P (2001) Seed dispersal by animals: exact identification of source trees with endocarp DNA microsatellites. Mol Ecol 10:2275–2283
Hall JB, O’Brien EM, Sinclair FL (2000) Prunus africana: a monograph. School of Agricultural and Forest Sciences Publication Number 18. University of Wales, Bangor, pp 104
Hall P, Chase MR, Bawa KS (1994) Low genetic-variation but high population differentiation in a common tropical forest tree species. Conserv Biol 8:471–482
Hamrick JL, Godt MJW (1997) Effects of life history traits on genetic diversity in plant species. In: Silverton J, Franco M, Harper JL (eds) Plant life histories: ecology, phylogeny and evolution. Cambridge Royal Society, Cambridge, pp 102–118
Hamrick JL, Murawski DA, Nason JD (1993) The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. Vegetatio 107/108:281–297
Jordano P, Godoy JA (2000) RAPD variation and population genetic structure in Prunus mahaleb (Rosaceae), an animal-dispersed tree. Mol Ecol 9:1293–1305
Kalinowski ST (2005) hp-rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
Kalkman C (1965) The Old World species of Prunus subg. Laurocerasus, including those formerly referred to Pygeum. J. J. Groen & Zoon, Leiden
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
KIFCON (1994) Kakamega Forest. The official guide. (KIFCON) Kenya Indigenous Forest Conservation Programme. Nairobi, Kenya
Knowles P (1991) Spatial genetic structure within two natural stands of black spruce (Picea mariana (Mill.) B.S.P). Silvae Genet 40:13–19
Kokwaro JO (1988) Conservation status of the Kakamega Forest in Kenya: the easternmost relict of the equatorial rain forests of Africa. Monographs in Systematic Botany from the Missouri Botanical Garden 25:471–489
Lowe AJ, Boshier D, Ward M, Bacles CFE, Navarro C (2005) Genetic resource impacts of habitat loss and degradation; reconciling empirical evidence and predicted theory for neotropical trees, Heredity 95:255–273
Luck GW, Daily GC, Ehrlich PR (2003) Population diversity and ecosystem services. Trends Ecol Evol 18:331–336
McLaughlin JF, Hellman JJ, Boggs CL, Ehrlich PR (2002) Climate change hastens population extinctions. Proc Natl Acad Sci USA 99:6070–6074
Miller MP (1997) Tools for populations genetics analyses (TFPGA) 1.3. a Windows program for the analysis of allozyme and molecular population genetic data. Computer software distributed by author; http: //bioweb.usu.edu/mpmbio/tfpga.htm
Mitchell N (2004) The exploitation, disturbance history of Kakamega Forest, western Kenya. In: Bleher B, Dalitz H (eds) BIOTA-East Report No 1, vol 20. Bielefelder Ökologische Beiträge, Bielefeld
Murawski DA, Hamrick JL (1992) The mating system of Cavanillesia platanifolia under extremes of flowering-tree density: a test of predictions. Biotropica 26:141–159
Nason JD, Hamrick JL (1997) Reproductive and genetic consequences of forest fragmentation: Two case studies of neotropical canopy trees. J Hered 88:264–276
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Pacheco LF, Simonetti JA (2000) Genetic structure of a mimosoid tree deprived of its seed disperser, the spider monkey. Conserv Biol 14:1766–1775
Perry DJ, Knowles P (1991) Spatial genetic structure within three sugar maple (Acer saccharum Marsh.) stands. Heredity 66:137–142
Prober SM, Brown AHD (1994) Conservation of the grassy white box Woodlands - population- genetics and fragmentation of Eucalyptus albens. Conserv Biol 8:1003–1013
Raymond M, Rousset F (1995a) Genepop (version 1.2), population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Raymond M, Rousset F (1995b) An exact test for population differentiation. Evolution 49:1280–1283
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Rossetto M, Jones R, Hunter J (2004) Genetic effects of rainforest fragmentation in an early successional tree (Elaeocarpus grandis). Heredity 93:610–618
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Shapcott A (1999) Vagility and the monsoon rain forest archipelago of northern Australia: Patterns of genetic diversity in Syzygium nervosum (Myrtaceae). Biotropica 31:579–590
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Sosinski B, Gannavarapu M, Hager LD et al (2000) Characterization of microsatellite markers in peach Prunus persica (L.) Batsch. Theor App Genet 101:421–428
Tsingalia MH (1990) Habitat disturbance, severity and patterns of abundance in Kakamega Forest, Western Kenya. Afr J Ecol 28 213–226
Ueno S, Tomaru N, Yoshimaru H, Manabe T, Yamamoto S (2000) Genetic structure of Camellia japonica L. in an old-growth evergreen forest, Tsushima, Japan. Mol Ecol 9:647–656
White GM, Boshier DH, Powell W (2002) Increased pollen flow counteracts fragmentation in a tropical dry forest: an example from Swietenia humilis Zuccarini. Proc Natl Acad Sci USA 99:2038–2042
Xie C-Y, Knowles P (1991) Genetic substructure within natural populations of jack pine (Pinus banksiana Lamb.). Can J Bot 69:547–551
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Acknowledgements
We thank the Kenyan Ministry for Education & Research for research permission, the KWS and FD for granting us access to their reserves, Jasper M. Kirika for help with field work and Bärbel Bleher for support. We are grateful to D. G. Berens, T. Diegisser, E. M. Griebeler, J. Johannesen, F. A. Voigt and two anonymous reviewers for comments on earlier drafts of the manuscript. Financial support was provided by the BMBF (Biota East Africa 01LC0025/01LC0405).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Farwig, N., Braun, C. & Böhning-Gaese, K. Human disturbance reduces genetic diversity of an endangered tropical tree, Prunus africana (Rosaceae). Conserv Genet 9, 317–326 (2008). https://doi.org/10.1007/s10592-007-9343-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-007-9343-x