Abstract
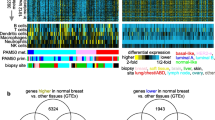
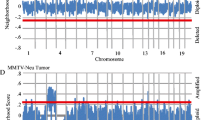
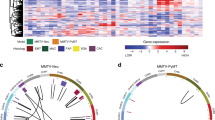
Microarray expression signature analyses have suggested that extracellular matrix (ECM) gene dysregulation is predictive of metastasis in both mouse mammary tumorigenesis and human breast cancer. We have previously demonstrated that such ECM dysregulation is influenced by hereditary germline-encoded variation. To identify novel metastasis efficiency modifiers, we performed expression QTL (eQTL) mapping in recombinant inbred mice by characterizing genetic loci modulating metastasis-predictive ECM gene expression. Three reproducible eQTLs were observed on chromosomes 7, 17 and 18. Candidate genes were identified by correlation analyses and known associations with metastasis. Seven candidates were identified (Ndn, Pi16, Luc7l, Rrp1b, Brd4, Centd3 and Csf1r). Stable transfection of the highly metastatic Mvt-1 mouse mammary tumor cell line with expression vectors encoding each candidate modulated metastasis-predictive ECM gene expression. Implantation of these cells into mice demonstrated that candidate gene ectopic expression impacts tumor progression. Gene expression analyses facilitated the construction of a transcriptional network that we have termed the ‘Diasporin Pathway’. This pathway contains the seven candidates, as well as metastasis-predictive ECM genes and metastasis suppressors. Brd4 and Rrp1b appear to form a central node within this network, which likely is a consequence of their physical interaction with the metastasis efficiency modifier Sipa1. Furthermore, we demonstrate that the microarray gene expression signatures induced by activation of ECM eQTL genes in the Mvt-1 cell line can be used to accurately predict survival in a human breast cancer cohort. These data imply that the Diasporin Pathway may be an important nexus in tumor progression in both mice and humans.




Similar content being viewed by others
References
Chung CT, Carlson RW (2003) Goals and objectives in the management of metastatic breast cancer. Oncologist 8:514–520
Guarneri V, Conte PF (2004) The curability of breast cancer and the treatment of advanced disease. Eur J Nucl Med Mol Imaging 31(Suppl 1):S149–S161
Ramaswamy S, Ross KN, Lander ES et al (2003) A molecular signature of metastasis in primary solid tumors. Nat Genet 33:49–54
van’t Veer LJ, Dai H, van de Vijver MJ et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536
van de Vijver MJ, He YD, van’t Veer LJ et al (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347:1999–2009
Weigelt B, Peterse JL, van’t Veer LJ (2005) Breast cancer metastasis: markers and models. Nat Rev Cancer 5:591–602
Lifsted T, Le Voyer T, Williams M et al (1998) Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int J Cancer 77:640–644
Guy CT, Cardiff RD, Muller WJ (1992) Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol 12:954–961
Park YG, Zhao X, Lesueur F et al (2005) Sipa1 is a candidate for underlying the metastasis efficiency modifier locus Mtes1. Nat Genet 37:1055–1062
Crawford NP, Qian X, Ziogas A et al (2007) Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLoS Genet 3:e214
Yang H, Rouse J, Lukes L et al (2004) Caffeine suppresses metastasis in a transgenic mouse model: a prototype molecule for prophylaxis of metastasis. Clin Exp Metastasis 21:719–735
Yang H, Crawford N, Lukes L et al (2005) metastasis predictive signature profiles pre-exist in normal tissues. Clin Exp Metastasis 22:593–603
Eckhardt BL, Parker BS, van Laar RK et al (2005) Genomic analysis of a spontaneous model of breast cancer metastasis to bone reveals a role for the extracellular matrix. Mol Cancer Res 3:1–13
Mucenski ML, Taylor BA, Jenkins NA et al (1986) AKXD recombinant inbred strains: models for studying the molecular genetic basis of murine lymphomas. Mol Cell Biol 6:4236–4243
Jang MK, Mochizuki K, Zhou M et al (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19:523–534
Farina A, Hattori M, Qin J et al (2004) Bromodomain protein Brd4 binds to GTPase-activating SPA-1, modulating its activity and subcellular localization. Mol Cell Biol 24:9059–9069
Dey A, Chitsaz F, Abbasi A et al (2003) The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A 100:8758–8763
Wang J, Williams RW, Manly KF (2003) WebQTL: web-based complex trait analysis. Neuroinformatics 1:299–308
Pei XF, Noble MS, Davoli MA et al (2004) Explant-cell culture of primary mammary tumors from MMTV-c-Myc transgenic mice. In vitro Cell Dev Biol Anim 40:14–21
Bailey D (1981) Recombinant inbred strains and bilineal congenic strains. In: Fox J (ed) The mouse in biomedical research, 1st edn. Academic, New York
Threadgill DW, Hunter KW, Zou F et al (2004) Genetic modifiers. In: Holland EC (ed) Mouse models of human cancer, 1st edn. Wiley-Liss, New York
Lancaster M, Rouse J, Hunter KW (2005) Modifiers of mammary tumor progression and metastasis on mouse chromosomes 7, 9, and 17. Mamm Genome 16:120–126
Schadt EE, Lamb J, Yang X et al (2005) An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet 37:710–717
Chesler EJ, Lu L, Shou S et al (2005) Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet 37:233–242
Maruyama K, Usami M, Aizawa T et al (1991) A novel brain-specific mRNA encoding nuclear protein (necdin) expressed in neurally differentiated embryonal carcinoma cells. Biochem Biophys Res Commun 178:291–296
Kimura E, Hidaka K, Kida Y et al (2004) Serine-arginine-rich nuclear protein Luc7l regulates myogenesis in mice. Gene 341:41–47
Reeves JR, Xuan JW, Arfanis K et al (2005) Identification, purification and characterization of a novel human blood protein with binding affinity for prostate secretory protein of 94 amino acids. Biochem J 385:105–114
Stacey TTI, Nie Z, Stewart A et al (2004) ARAP3 is transiently tyrosine phosphorylated in cells attaching to fibronectin and inhibits cell spreading in a RhoGAP-dependent manner. J Cell Sci 117:6071–6084
Rohde CM, Schrum J, Lee AW (2004) A juxtamembrane tyrosine in the colony stimulating factor-1 receptor regulates ligand-induced Src association, receptor kinase function, and down-regulation. J Biol Chem 279:43448–43461
Hu B, Wang S, Zhang Y et al (2003) A nuclear target for interleukin-1alpha: interaction with the growth suppressor necdin modulates proliferation and collagen expression. Proc Natl Acad Sci U S A 100:10008–10013
Moon HE, Ahn MY, Park JA et al (2005) Negative regulation of hypoxia inducible factor-1alpha by necdin. FEBS Lett 579:3797–3801
Semenza GL (2000) Hypoxia, clonal selection, and the role of HIF-1 in tumor progression. Crit Rev Biochem Mol Biol 35:71–103
Wrobel CN, Debnath J, Lin E et al (2004) Autocrine CSF-1R activation promotes Src-dependent disruption of mammary epithelial architecture. J Cell Biol 165:263–273
Lin EY, Nguyen AV, Russell RG et al (2001) Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med 193:727–740
Jackson P, Marreiros A, Russell PJ (2005) KAI1 tetraspanin and metastasis suppressor. Int J Biochem Cell Biol 37:530–534
Amirkhosravi A, Meyer T, Chang JY et al (2002) Tissue factor pathway inhibitor reduces experimental lung metastasis of B16 melanoma. Thromb Haemost 87:930–936
Salerno M, Ouatas T, Palmieri D et al (2003) Inhibition of signal transduction by the nm23 metastasis suppressor: possible mechanisms. Clin Exp Metastasis 20:3–10
Liu WM, Zhang XA (2006) KAI1/CD82, a tumor metastasis suppressor. Cancer Lett 240:183–194
Crawford NP, Ziogas A, Peel DJ et al (2006) Polymorphisms of SIPA1 are associated with metastasis and other indicators of poor prognosis in breast cancer. Breast Cancer Res 8:R16
Itoh M, Nelson CM, Myers CA et al (2007) Rap1 integrates tissue polarity, lumen formation, and tumorigenic potential in human breast epithelial cells. Cancer Res 67:4759–4766
Retta SF, Balzac F, Avolio M (2006) Rap1: a turnabout for the crosstalk between cadherins and integrins. Eur J Cell Biol 85:283–293
Bild AH, Yao G, Chang JT et al (2006) Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 439:353–357
Bogaerts J, Cardoso F, Buyse M et al (2006) Gene signature evaluation as a prognostic tool: challenges in the design of the MINDACT trial. Nat Clin Pract Oncol 3:540–551
Acknowledgements
We thank Drs. Doug Lowy and Jude Alsarraj for critical comments on this manuscript. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Crawford, N.P.S., Walker, R.C., Lukes, L. et al. The Diasporin Pathway: a tumor progression-related transcriptional network that predicts breast cancer survival. Clin Exp Metastasis 25, 357–369 (2008). https://doi.org/10.1007/s10585-008-9146-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-008-9146-6