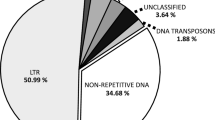
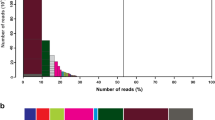
Abstract
Structural karyotype changes result from ectopic recombination events frequently associated with repetitive DNA. Although most Phaseolus species present relatively stable karyotypes with 2n = 22 chromosomes, the karyotypes of species of the Leptostachyus group show high rates of structural rearrangements, including a nested chromosome fusion that led to the dysploid chromosome number of the group (2n = 20). We examined the roles of repetitive landscapes in the rearrangements of species of the Leptostachyus group using genome-skimming data to characterize the repeatome in a range of Phaseolus species and compared them to species of that group (P. leptostachyus and P. macvaughii). LTR retrotransposons, especially the Ty3/gypsy lineage Chromovirus, were the most abundant elements in the genomes. Differences in the abundance of Tekay, Retand, and SIRE elements between P. macvaughii and P. leptostachyus were reflected in their total amounts of Ty3/gypsy and Ty1/copia. The satellite DNA fraction was the most divergent among the species, varying both in abundance and distribution, even between P. leptostachyus and P. macvaughii. The rapid turnover of repeats in the Leptostachyus group may be associated with the several rearrangements observed.



Similar content being viewed by others
Data availability
Sequencing data generated during this study are available on the NCBI SRA platform (BioProject number PRJNA865793) under the following accession numbers: SAMN30133279 (Phaseolus leptostachyus), SAMN30133280 (P. macvaughii), and SAMN30133281 (P. lunatus). New satellite DNA sequences are available under the following accession numbers: BankIt2739180 OR500523 to OR500540.
Code availability
Not applicable.
Abbreviations
- CD:
-
Chromodomain
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- dNTP:
-
Deoxynucleotide triphosphates
- FISH:
-
Fluorescence in situ hybridization
- LINE:
-
Long interspersed nuclear elements
- LTR:
-
Long terminal repeats
- Mya:
-
Million years ago
- NCF:
-
Nested chromosome fusion
- PCR:
-
Polymerase chain reaction
- PTD:
-
Putative target domains
- satDNA:
-
Satellite DNA
- SINE:
-
Short interspersed nuclear elements
- SSC:
-
Saline-sodium citrate
- TAREAN:
-
TAndem REpeat ANalyzer
- TBT:
-
Trehalose, bovine serum albumin (BSA), and polysorbate-20 (Tween-20)
- TNT:
-
Tree analysis using New Technology
References
Almeida C, Pedrosa-Harand A (2013) High macro-collinearity between lima bean (Phaseolus lunatus L.) and the common bean (P. vulgaris L.) as revealed by comparative cytogenetic mapping. Theor Appl Genet 126:1909–1916. https://doi.org/10.1007/s00122-013-2106-9
Ayres-Alves T, Cardoso AL, Nagamachi CY et al (2017) Karyotypic evolution and chromosomal organization of repetitive DNA sequences in species of Panaque, Panaqolus, and Scobinancistrus (Siluriformes and Loricariidae) from the Amazon Basin. Zebrafish 14:251–260. https://doi.org/10.1089/zeb.2016.1373
Báez M, Vaio M, Dreissig S et al (2019) Together but different: the subgenomes of the bimodal Eleutherine karyotypes are differentially organized. Front Plant Sci 10:1–14. https://doi.org/10.3389/fpls.2019.01170
Biscotti MA, Olmo E, Heslop-Harrison JS, (Pat), (2015) Repetitive DNA in eukaryotic genomes. Chromosome Res 23:415–420. https://doi.org/10.1007/s10577-015-9499-z
Bourque G, Burns KH, Gehring M et al (2018) Ten things you should know about transposable elements. Genome Biol 19:199. https://doi.org/10.1186/s13059-018-1577-z
Broughton WJ, Hernández G, Blair M et al (2003) Beans (Phaseolus spp.) - model food legumes. Plant Soil 252:55–128
Cermak T, Kubat Z, Hobza R et al (2008) Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Res 16:961–976. https://doi.org/10.1007/s10577-008-1254-2
Chen NWG, Thareau V, Ribeiro T et al (2018) Common bean subtelomeres are hot spots of recombination and favor resistance gene evolution. Front Plant Sci 9:1–15. https://doi.org/10.3389/fpls.2018.01185
David P, Chen NWG, Pedrosa-Harand A et al (2009) A nomadic subtelomeric disease resistance gene cluster in common bean. Plant Physiol 151:1048–1065. https://doi.org/10.1104/pp.109.142109
Delgado-Salinas A, Bibler R, Lavin M (2006) Phylogeny of the genus Phaseolus (Leguminosae): a recent diversification in an ancient landscape. Syst Bot 31:779–791. https://doi.org/10.1600/036364406779695960
Dodsworth S, Chase MW, Kelly LJ et al (2015) Genomic repeat abundances contain phylogenetic signal. Syst Biol 64:112–126. https://doi.org/10.1093/sysbio/syu080
Dodsworth S, Jang TS, Struebig M et al (2017) Genome-wide repeat dynamics reflect phylogenetic distance in closely related allotetraploid Nicotiana (Solanaceae). Plant Syst Evol 303:1013–1020. https://doi.org/10.1007/s00606-016-1356-9
Doležel J, Sgorbati S, Lucretti S (1992) Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol Plant 85:625–631. https://doi.org/10.1111/j.1399-3054.1992.tb04764.x
Doležel J, Greilhuber J, Suda J (2007) Estimation of nuclear DNA content in plants using flow cytometry. Nat Protoc 2:2233–2244. https://doi.org/10.1038/nprot.2007.310
Ferraz ME, Fonsêca A, Pedrosa-Harand A (2020) Multiple and independent rearrangements revealed by comparative cytogenetic mapping in the dysploid Leptostachyus group (Phaseolus L., Leguminosae). Chromosome Res 28:395–405. https://doi.org/10.1007/s10577-020-09644-z
Fonsêca A, Ferraz ME, Pedrosa-Harand A (2016) Speeding up chromosome evolution in Phaseolus: multiple rearrangements associated with a one-step descending dysploidy. Chromosoma 125:413–421. https://doi.org/10.1007/s00412-015-0548-3
Fonsêca A, Pedrosa-Harand A (2017) Cytogenetics and comparative analysis of Phaseolus species. In: Pérez de la Vega M, Santalla M, Marsolais F (eds) The common bean genome. Compendium of plant genomes. Springer, Cham, pp 57–68. https://doi.org/10.1007/978-3-319-63526-2_3
Fransz P, Linc G, Lee CR et al (2016) Molecular, genetic and evolutionary analysis of a paracentric inversion in Arabidopsis thaliana. Plant J 88:159–178. https://doi.org/10.1111/tpj.13262
Gaiero P, Vaio M, Peters SA et al (2019) Comparative analysis of repetitive sequences among species from the potato and the tomato clades. Ann Bot 123:521–532. https://doi.org/10.1093/aob/mcy186
Gao D, Abernathy B, Rohksar D et al (2014) Annotation and sequence diversity of transposable elements in common bean (Phaseolus vulgaris). 5:1–9. https://doi.org/10.3389/fpls.2014.00339
Garcia T, Duitama J, Zullo S et al (2020) Comprehensive genomic resources related to domestication and crop improvement traits in Lima bean. Nat Res. https://doi.org/10.21203/rs.3.rs-95762/v1
Garrido-Ramos MA (2015) Satellite DNA in plants: more than just rubbish. Cytogenet Genome Res 146:153–170. https://doi.org/10.1159/000437008
Garrido-Ramos M (2017) Satellite DNA: an Evolving Topic. Genes (basel) 8:230. https://doi.org/10.3390/genes8090230
Gong Z, Wu Y, Koblížková A et al (2012) Repeatless and repeat-based centromeres in potato: Implications for centromere evolution. Plant Cell 24:3559–3574. https://doi.org/10.1105/tpc.112.100511
González ML, Chiapella JO, Urdampilleta JD (2018) Characterization of some satellite DNA families in Deschampsia antarctica (Poaceae). Polar Biol 41:457–468. https://doi.org/10.1007/s00300-017-2205-1
Heslop-Harrison JS, Harrison GE, Leitch IJ (1992) Reprobing of DNA: DNA in situ hybridization preparations. Trends Genet 8:372–373
Ishii T, Juranić M, Maheshwari S et al (2020) Unequal contribution of two paralogous CENH3 variants in cowpea centromere function. Commun Biol 3:775. https://doi.org/10.1038/s42003-020-01507-x
Iwata A, Tek AL, Richard MMS et al (2013) Identification and characterization of functional centromeres of the common bean. Plant J 76:47–60. https://doi.org/10.1111/tpj.12269
Iwata-Otsubo A, Lin JY, Gill N, Jackson SA (2016a) Highly distinct chromosomal structures in cowpea (Vigna unguiculata), as revealed by molecular cytogenetic analysis. Chromosome Res 24:197–216. https://doi.org/10.1007/s10577-015-9515-3
Iwata-Otsubo A, Radke B, Findley S et al (2016b) Fluorescence in situ hybridization (FISH)-based karyotyping reveals rapid evolution of centromeric and subtelomeric repeats in common bean (Phaseolus vulgaris) and relatives. G3 Genes. Genomes, Genet 6:1013–1022. https://doi.org/10.1534/g3.115.024984
Khost DE, Eickbush DG, Larracuente AM (2017) Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res 27:709–721. https://doi.org/10.1101/gr.213512.116
Koukalova B, Moraes AP, Renny-Byfield S et al (2010) Fall and rise of satellite repeats in allopolyploids of Nicotiana over c. 5 million years. New Phytol 186:148–160. https://doi.org/10.1111/j.1469-8137.2009.03101.x
Lambowitz AM, Gellert M, Chandler M, Craig NL, Sandmeyer SB, Rice PA (eds) (2015) Mobile DNA III. American Society of Microbiology
Li SF, Su T, Cheng GQ et al (2017) Chromosome evolution in connection with repetitive sequences and epigenetics in plants. Genes (Basel) 8. https://doi.org/10.3390/genes8100290
Lim KY, Skalicka K, Koukalova B et al (2004) Dynamic changes in the distribution of a satellite homologous to intergenic 26–18S rDNA spacer in the evolution of Nicotiana. Genetics 166:1935–1946. https://doi.org/10.1534/genetics.166.4.1935
Loureiro J, Rodriguez E, Dolezel J, Santos C (2007) Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot 100:875–888. https://doi.org/10.1093/aob/mcm152
Macas J, Novak P, Pellicer J et al (2015) In depth characterization of repetitive DNA in 23 plant genomes reveals sources of genome size variation in the legume tribe Fabeae. PLoS One 10. https://doi.org/10.1371/journal.pone.0143424
Mata-Sucre Y, Sader M, Van-Lume B et al (2020) How diverse is heterochromatin in the Caesalpinia group? Cytogenomic characterization of Erythrostemon hughesii Gagnon & G.P. Lewis (Leguminosae: Caesalpinioideae). Planta 252:49. https://doi.org/10.1007/s00425-020-03453-8
Maumus F, Quesneville H (2016) Impact and insights from ancient repetitive elements in plant genomes. Curr Opin Plant Biol 30:41–46. https://doi.org/10.1016/j.pbi.2016.01.003
Mehrotra S, Goyal V (2014) Repetitive sequences in plant nuclear DNA: types, distribution, evolution and function. Genomics Proteomics Bioinformatics 12:164–171. https://doi.org/10.1016/j.gpb.2014.07.003
Mercado-Ruaro P, Delgado-Salinas A (1998) Karyotypic studies on species of Phaseolus (Fabaceae: Phaseolinae). Am J Bot 85:1–9. https://doi.org/10.2307/2446547
Mercado-Ruaro P, Delgado-Salinas A (2000) Cytogenetic studies in Phaseolus L. (Fabaceae). Genet Mol Biol 23:985–987. https://doi.org/10.1590/S1415-47572000000400043
Mlinarec J, Franjević D, Harapin J, Besendorfer V (2016) The impact of the Tekay chromoviral elements on genome organisation and evolution of Anemone s.l. (Ranunculaceae). Plant Biol 18:332–347. https://doi.org/10.1111/plb.12393
Moghaddam SM, Oladzad A, Koh C et al (2021) The tepary bean genome provides insight into evolution and domestication under heat stress. Nat Commun 12:1–14. https://doi.org/10.1038/s41467-021-22858-x
Neumann P, Novák P, Hoštáková N, MacAs J (2019) Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mobile DNA 10:1. https://doi.org/10.1186/s13100-018-0144-1
Neumann P, Navrátilová A, Koblížková A et al (2011) Plant centromeric retrotransposons: a structural and cytogenetic perspective. Mob. DNA 2
Novák P, Neumann P, Macas J (2010) Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11:1–12. https://doi.org/10.1186/1471-2105-11-378
Novák P, Neumann P, Pech J et al (2013) RepeatExplorer: a galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 29:792–793. https://doi.org/10.1093/bioinformatics/btt054
Novák P, Robledillo LÁ, Koblížková A et al (2017) TAREAN: a computational tool for identification and characterization of satellite DNA from unassembled short reads. Nucleic Acids Res 45:1–10. https://doi.org/10.1093/nar/gkx257
Novák P, Neumann P, Macas J (2020) Global analysis of repetitive DNA from unassembled sequence reads using RepeatExplorer2. Nat Protoc. https://doi.org/10.1038/s41596-020-0400-y
Pedrosa A, Sandal N, Stougaard J et al (2002) Chromosomal map of the model legume Lotus japonicus. Genetics 161:1661–1672
Piednoël M, Carrete-Vega G, Renner SS (2013) Characterization of the LTR retrotransposon repertoire of a plant clade of six diploid and one tetraploid species. Plant J 75:699–709. https://doi.org/10.1111/tpj.12233
Pita S, Panzera F, Mora P et al (2017) Comparative repeatome analysis on Triatoma infestans Andean and Non-Andean lineages, main vector of Chagas disease. PLoS ONE 12:1–13. https://doi.org/10.1371/journal.pone.0181635
Prakhongcheep O, Thapana W, Suntronpong A et al (2017) Lack of satellite DNA species-specific homogenization and relationship to chromosomal rearrangements in monitor lizards (Varanidae, Squamata). BMC Evol Biol 17:1–14. https://doi.org/10.1186/s12862-017-1044-6
Puterova J, Kubat Z, Kejnovsky E et al (2018) The slowdown of Y chromosome expansion in dioecious Silene latifolia due to DNA loss and male-specific silencing of retrotransposons. BMC Genomics 19. https://doi.org/10.1186/s12864-018-4547-7
Raskina O, Belyayev A, Nevo E (2004) Activity of the En/Spm-like transposons in meiosis as a base for chromosome repatterning in a small, isolated, peripheral population of Aegilops speltoides Tausch. Chromosome Res 12:153–161
Rendón-Anaya M, Montero-Vargas JM, Saburido-Álvarez S et al (2017) Genomic history of the origin and domestication of common bean unveils its closest sister species. Genome Biol 18:1–17. https://doi.org/10.1186/s13059-017-1190-6
Ribeiro T, Marques A, Novák P, Schubert V, Vanzela AL, Macas J, Houben A, Pedrosa-Harand A (2017a) Centromeric and non-centromeric satellite DNA organisation differs in holocentric Rhynchospora species. Chromosoma 126(2):325–335. https://doi.org/10.1007/s00412-016-0616-3
Ribeiro T, dos Santos KGB, Richard MMS et al (2017b) Evolutionary dynamics of satellite DNA repeats from Phaseolus beans. Protoplasma 254:791–801. https://doi.org/10.1007/s00709-016-0993-8
Ribeiro T, Vasconcelos E, dos Santos KGB et al (2020) Diversity of repetitive sequences within compact genomes of Phaseolus L. beans and allied genera Cajanus L. and Vigna Savi. Chromosome Res 28:139–153. https://doi.org/10.1007/s10577-019-09618-w
Richard MMS, Chen NWG, Thareau V et al (2013) The subtelomeric khipu satellite repeat from Phaseolus vulgaris: lessons learned from the genome analysis of the andean genotype G19833. Front Plant Sci 4:1–15. https://doi.org/10.3389/fpls.2013.00109
Robledillo ÁL, Koblížková A, Novák P et al (2018) Satellite DNA in Vicia faba is characterized by remarkable diversity in its sequence composition, association with centromeres, and replication timing. Sci Rep 8:5838. https://doi.org/10.1038/s41598-018-24196-3
Ruiz-Ruano FJ, López-León MD, Cabrero J, Camacho JPM (2016) High-throughput analysis of the satellitome illuminates satellite DNA evolution. Sci Rep 6:1–14. https://doi.org/10.1038/srep28333
Sader M, Vaio M, Cauz-Santos LA et al (2021) Large vs small genomes in Passiflora: the influence of the mobilome and the satellitome. Planta 253:1–18. https://doi.org/10.1007/s00425-021-03598-0
Schmutz J, McClean PE, Mamidi S et al (2014) A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet 46:707–713. https://doi.org/10.1038/ng.3008
Schubert I, Lysak MA (2011) Interpretation of karyotype evolution should consider chromosome structural constraints. Trends Genet 27:207–216
Siljak-Yakovlev S, Godelle B, Zoldos V et al (2017) Evolutionary implications of heterochromatin and rDNA in chromosome number and genome size changes during dysploidy: a case study in Reichardia genus. PLoS ONE 12:1–21. https://doi.org/10.1371/journal.pone.0182318
Smalec BM, Heider TN, Flynn BL, O’Neill RJ (2019) A centromere satellite concomitant with extensive karyotypic diversity across the Peromyscus genus defies predictions of molecular drive. Chromosome Res 27:237–252. https://doi.org/10.1007/s10577-019-09605-1
Sonnhammer ELL, Durbin R (1995) A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene 167:GC1–GC10. https://doi.org/10.1016/0378-1119(95)00714-8
Van-Lume B, Mata-Sucre Y, Báez M et al (2019) Evolutionary convergence or homology? Comparative cytogenomics of Caesalpinia group species (Leguminosae) reveals diversification in the pericentromeric heterochromatic composition. Planta 250:2173–2186. https://doi.org/10.1007/s00425-019-03287-z
Vlasova A, Capella-Gutiérrez S, Rendón-Anaya M et al (2016) Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol 17:1–18. https://doi.org/10.1186/s13059-016-0883-6
Vondrak T, Ávila Robledillo L, Novák P et al (2020) Characterization of repeat arrays in ultra-long nanopore reads reveals frequent origin of satellite DNA from retrotransposon-derived tandem repeats. Plant J 101:484–500. https://doi.org/10.1111/tpj.14546
Weising K, Nybom H, Wolff K, Kahl G (2005) CTAB protocol I. In: Taylor & Francis Group (ed) DNA fingerprinting in plants : principles, methods, and applications, 2nd edn. Boca Raton, pp 100–102
Wessler SR (2006) Eukaryotic transposable elements: teaching old genomes new tricks. In: Caporale LH (ed) The Implicit Genome. Oxford University Press, New York, pp 138–165
Zhang H, Koblížková A, Wang K et al (2014) Boom-bust turnovers of megabase-sized centromeric DNA in Solanum species: rapid evolution of DNA sequences associated with centromeres. Plant Cell 26:1436–1447. https://doi.org/10.1105/tpc.114.123877
Zhu Q, Cai Z, Tang Q, Jin W (2016) Repetitive sequence analysis and karyotyping reveal different genome evolution and speciation of diploid and tetraploid Tripsacum dactyloides. Crop J 4:247–255. https://doi.org/10.1016/j.cj.2016.04.003
Acknowledgements
The authors are grateful to EMBRAPA CENARGEN and EMBRAPA Arroz e Feijão (Brazil), CIAT (Colômbia), and IPK Gatersleben (Germany), for providing seed material. We are also grateful to CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brazil) for financial support.
Funding
This study was supported by CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil) through a fellowship awarded to A. Pedrosa-Harand (312694/2021–0) and was supported in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brazil (CAPES, Finance Code 001).
Author information
Authors and Affiliations
Contributions
MEF performed experiments, and drafted the manuscript. TR and MAS performed bioinformatic analyses and discussed the data. TN performed experiments and bioinformatic analyses and organized figures. APH designed and supervised the research and corrected the manuscript. All authors read, discussed, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consented for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Rachel O'Neill
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key message
The bioinformatic and cytogenetic characterization of the repeatome of 13 accessions of 11 Phaseolus species revealed rapid turnovers and distribution changes in some LTR-retrotransposon lineages and satellite DNA families in the Leptostachyus group, which experienced extensive rearrangements after a dysploidy event.
Supplementary Information
Below is the link to the electronic supplementary material.
10577_2023_9739_MOESM1_ESM.tif
Supplementary Figure 1. New genome size estimates of Phaseolus species by flow cytometry. A) P. microcarpus; B) P. dumosus. Solanum lycopersicum var. Stupicke was used as an internal standard. (TIF 3774 KB)
10577_2023_9739_MOESM2_ESM.tif
Supplementary Figure 2. Dotplot analysis of newly identified and shared satDNAs from individual analyses of P. macvaughii, P. leptostachyus and P. lunatus using TAREAN, compared to khipu. (TIF 5901 KB)
10577_2023_9739_MOESM3_ESM.tif
Supplementary Figure 3. Monomer sequence alignments of newly identified and shared satDNAs from individual analyses of P. macvaughii, P. leptostachyus and P. lunatus using TAREAN, compared to khipu. A) PleSAT592 and PmaSAT647; B) khipu consensus from P. vulgaris compared to khipu variants with 548 bp, 549 bp and 558 bp; C) PleSAT572 and PluSAT940 compared to PmiSAT30. (TIF 19262 KB)
10577_2023_9739_MOESM4_ESM.tif
Supplementary Figure 4. Complex organization of PluSAT940 with insertions of PmiSAT30-like repeats. The percentage values represent the similarity among the variations of PmiSAT30 sequence with the monomer consensus. (TIF 10324 KB)
10577_2023_9739_MOESM5_ESM.tif
Supplementary Figure 5. Chromosomal localization of satDNA families in P. macvaughii showing very weak dispersed signals. (a-b), PleSAT752, (c-d) khipu. Bar in d = 5 µm. (TIF 3103 KB)
10577_2023_9739_MOESM7_ESM.docx
Supplementary file 7. Supplementary Data 1. Monomers consensus sequences of satDNAs identified either in individual or comparative clustering analysis in Phaseolus species using Repeat Explorer. (DOCX 23 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ferraz, M.E., Ribeiro, T., Sader, M. et al. Comparative analysis of repetitive DNA in dysploid and non-dysploid Phaseolus beans. Chromosome Res 31, 30 (2023). https://doi.org/10.1007/s10577-023-09739-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10577-023-09739-3