Abstract
Telomerase is a ribonucleoprotein ribonucleic enzyme that elongates telomere repeat sequences at the ends of chromosomes and contributes to cellular immortalization. The catalytic component of telomerase, human telomerase reverse transcriptase (hTERT), has been observed to be reactivated in immortalized cells. Notably, most cancer cells have been found to have active hTERT mRNA transcription, resulting in continuous cell division, which is crucial for malignant transformation. Therefore, discovering mechanisms underlying the regulation of hTERT transcription is an attractive target for cancer-specific treatments.
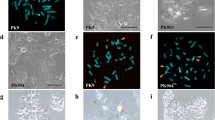
Loss of heterozygosity (LOH) of chromosome 3p21.3 has been frequently observed in human oral squamous cell carcinoma (OSCC). Moreover, we previously reported that HSC3 OSCC microcell hybrid clones with an introduced human chromosome 3 (HSC3#3) showed inhibition of hTERT transcription compared with the parental HSC3 cells. This study examined whether hTERT transcription regulators are present in the 3p21.3 region. We constructed a human artificial chromosome (HAC) vector (3p21.3-HAC) with only the 3p21.3-p22.2 region and performed functional analysis using the 3p21.3-HAC. HSC3 microcell hybrid clones with an introduced 3p21.3-HAC exhibited significant suppression of hTERT transcription, similar to the microcell hybrid clones with an intact chromosome 3. In contrast, HSC3 clones with truncated chromosome 3 with deletion of the 3p21.3 region (3delp21.3) showed no effect on hTERT expression levels. These results provide direct evidence that hTERT suppressor gene(s) were retained in the 3p21.3 region, suggesting that the presence of regulatory factors that control telomerase enzyme activity may be involved in the development of OSCC.






Similar content being viewed by others
Data availability
The datasets created and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Change history
17 August 2023
A Correction to this paper has been published: https://doi.org/10.1007/s10577-023-09734-8
Abbreviations
- CYP3A :
-
Cytochrome P450 family 3 subfamily A
- DMD :
-
Duchenne muscular dystrophy
- DYS-HAC :
-
HAC vectors containing the entire dystrophin genomic region
- FISH :
-
Fluorescence in situ hybridization
- HAC :
-
Human artificial chromosome
- hTERT :
-
Human telomerase reverse transcriptase
- LOH :
-
Loss of heterozygosity
- MMCT :
-
Microcell-mediated chromosome transfer
- OSCC :
-
Oral squamous cell carcinoma
- RCC :
-
Renal cell carcinoma
- STS :
-
Sequence-tagged site
- 3p21.3-HAC :
-
HAC vector with only the 3p21.3-p22.2 region
- 3delp21.3 :
-
Truncated chromosome 3 with deletion of the 3p21.3 region
References
Abe S, Tanaka H, Notsu T, Horike S, Fujisaki C, Qi DL, Ohhira T, Gilley D, Oshimura M, Kugoh H (2010) Localization of an hTERT repressor region on human chromosome 3p21.3 using chromosome engineering. Genome Integr 1(1):6. https://doi.org/10.1186/2041-9414-1-6
Bernardes de Jesus B, Blasco MA (2013) Telomerase at the intersection of cancer and aging. Trends Genet 29(9):513–520. https://doi.org/10.1016/j.tig.2013.06.007
Bhan A, Soleimani M, Mandal SS (2017) Long noncoding RNA and cancer: a new paradigm. Cancer Res 77(15):3965–3981. https://doi.org/10.1158/0008-5472.CAN-16-2634
Chang JW, Hsu HS, Ni HJ, Chuang CT, Hsiung CH, Huang TH, Wang YC (2010) Distinct epigenetic domains separated by a CTCF bound insulator between the tandem genes, BLU and RASSF1A. PLOS ONE 5(9):e12847. https://doi.org/10.1371/journal.pone.0012847
Dixon JR, Gorkin DU, Ren B (2016) Chromatin domains: The unit of chromosome organization. Mol Cell 62(5):668–680. https://doi.org/10.1016/j.molcel.2016.05.018
Gissi DB, Morandi L, Gabusi A, Tarsitano A, Marchetti C, Cura F, Palmieri A, Montebugnoli L, Asioli S, Foschini MP, Scapoli L (2018) A noninvasive test for microRNA expression in oral squamous cell carcinoma. Int J Mol Sci 19(6). https://doi.org/10.3390/ijms19061789
Hesson LB, Cooper WN, Latif F (2007) Evaluation of the 3p21.3 tumour-suppressor gene cluster. Oncogene 26(52):7283–7301. https://doi.org/10.1038/sj.onc.1210547
Jin S, Yang X, Li J, Yang W, Ma H, Zhang Z (2019) p53-targeted lincRNA-p21 acts as a tumor suppressor by inhibiting JAK2/STAT3 signaling pathways in head and neck squamous cell carcinoma. Mol Cancer 18(1):38. https://doi.org/10.1186/s12943-019-0993-3
Kasamatsu A, Uzawa K, Usukura K, Koike K, Nakashima D, Ishigami T, Fushimi K, Ogawara K, Shiiba M, Tanzawa H (2011) Loss of heterozygosity in oral cancer. Oral Sci Int 8(2):37–43. https://doi.org/10.1016/S1348-8643(11)00027-9
Katoh M, Ayabe F, Norikane S, Okada T, Masumoto H, Horike S, Shirayoshi Y, Oshimura M (2004) Construction of a novel human artificial chromosome vector for gene delivery. Biochem Biophys Res Commun 321(2):280–290. https://doi.org/10.1016/j.bbrc.2004.06.145
Kazuki Y, Oshimura M (2011) Human artificial chromosomes for gene delivery and the development of animal models. Mol Ther 19(9):1591–1601. https://doi.org/10.1038/mt.2011.136
Kazuki Y, Hiratsuka M, Takiguchi M, Osaki M, Kajitani N, Hoshiya H, Hiramatsu K, Yoshino T, Kazuki K, Ishihara C, Takehara S, Higaki K, Nakagawa M, Takahashi K, Yamanaka S, Oshimura M (2010) Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol Ther 18(2):386–393. https://doi.org/10.1038/mt.2009.274
Kazuki Y, Hoshiya H, Takiguchi M, Abe S, Iida Y, Osaki M, Katoh M, Hiratsuka M, Shirayoshi Y, Hiramatsu K, Ueno E, Kajitani N, Yoshino T, Kazuki K, Ishihara C, Takehara S, Tsuji S, Ejima F, Toyoda A, Sakaki Y, Larionov V, Kouprina N, Oshimura M (2011) Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther 18(4):384–393. https://doi.org/10.1038/gt.2010.147
Kazuki Y, Kobayashi K, Aueviriyavit S, Oshima T, Kuroiwa Y, Tsukazaki Y, Senda N, Kawakami H, Ohtsuki S, Abe S, Takiguchi M, Hoshiya H, Kajitani N, Takehara S, Kubo K, Terasaki T, Chiba K, Tomizuka K, Oshimura M (2013) Trans-chromosomic mice containing a human CYP3A cluster for prediction of xenobiotic metabolism in humans. Hum Mol Genet 22(3):578–592. https://doi.org/10.1093/hmg/dds468
Kempfer R, Pombo A (2020) Methods for mapping 3D chromosome architecture. Nat Rev Genet 21(4):207–226. https://doi.org/10.1038/s41576-019-0195-2
Kononenko AV, Lee NC, Earnshaw WC, Kouprina N, Larionov V (2013) Re-engineering an alphoid(tetO)-HAC-based vector to enable high-throughput analyses of gene function. Nucleic Acids Res 41(10):e107. https://doi.org/10.1093/nar/gkt205
Kugoh H, Ohira T, Oshimura M (2015) Studies of tumor suppressor genes via chromosome engineering. Cancers (Basel) 8(1). https://doi.org/10.3390/cancers8010004
Léveillé N, Melo CA, Rooijers K, Díaz-Lagares A, Melo SA, Korkmaz G, Lopes R, Moqadam FA, Maia AR, Wijchers PJ, Geeven G, den Boer ML, Kalluri R, de Laat W, Esteller M, Agami R (2015) Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun 6:6520. https://doi.org/10.1038/ncomms7520
Liao A, Tan G, Chen L, Zhou W, Hu H (2016) RASSF1A inhibits gastric cancer cell proliferation by miR-711- mediated downregulation of CDK4 expression. Oncotarget 7(5):5842–5851. https://doi.org/10.18632/oncotarget.6813
Melo CA, Drost J, Wijchers PJ, van de Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Léveillé N, Kalluri R, de Laat W, Agami R (2013) eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49(3):524–535. https://doi.org/10.1016/j.molcel.2012.11.021
Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima-Abo A, Kotani K, Oikawa H, Sakurai E, Izutsu N, Kato K, Komatsu H, Ikeda K, Wakabayashi G, Masuda T (2008) Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci 99(2):280–286. https://doi.org/10.1111/j.1349-7006.2007.00666.x
Nishio S, Ohira T, Sunamura N, Oshimura M, Ryoke K, Kugoh H (2015) Repression of hTERT transcription by the introduction of chromosome 3 into human oral squamous cell carcinoma. Biochem Biophys Res Commun 466(4):755–759. https://doi.org/10.1016/j.bbrc.2015.09.119
Ohira T, Naohiro S, Nakayama Y, Osaki M, Okada F, Oshimura M, Kugoh H (2015) miR-19b regulates hTERT mRNA expression through targeting PITX1 mRNA in melanoma cells. Sci Rep 5:8201. https://doi.org/10.1038/srep08201
Ohira T, Kojima H, Kuroda Y, Aoki S, Inaoka D, Osaki M, Wanibuchi H, Okada F, Oshimura M, Kugoh H (2019a) PITX1 protein interacts with ZCCHC10 to regulate hTERT mRNA transcription. PLOS ONE 14(8):e0217605. https://doi.org/10.1371/journal.pone.0217605
Ohira T, Miyauchi K, Uno N, Shimizu N, Kazuki Y, Oshimura M, Kugoh H (2019b) An efficient protein production system via gene amplification on a human artificial chromosome and the chromosome transfer to CHO cells. Sci Rep 9(1):16954. https://doi.org/10.1038/s41598-019-53116-2
Peng WX, Koirala P, Mo YY (2017) LncRNA-mediated regulation of cell signaling in cancer. Oncogene 36(41):5661–5667. https://doi.org/10.1038/onc.2017.184
Piao CQ, Liu L, Zhao YL, Balajee AS, Suzuki M, Hei TK (2005) Immortalization of human small airway epithelial cells by ectopic expression of telomerase. Carcinogenesis 26(4):725–731. https://doi.org/10.1093/carcin/bgi016
Ransohoff JD, Wei Y, Khavari PA (2018) The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol 19(3):143–157. https://doi.org/10.1038/nrm.2017.104
Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA (2002) The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol 22(12):4309–4318. https://doi.org/10.1128/MCB.22.12.4309-4318.2002
Tanaka H, Shimizu M, Horikawa I, Kugoh H, Yokota J, Barrett JC, Oshimura M (1998) Evidence for a putative telomerase repressor gene in the 3p142-p211 region. Genes Chromosomes Cancer 23(2):123–133. https://doi.org/10.1002/(sici)1098-2264(199810)23:2%3c123::aid-gcc5%3e3.0.co;2-4
Tommasi S, Dammann R, Zhang Z, Wang Y, Liu L, Tsark WM, Wilczynski SP, Li J, You M, Pfeifer GP (2005) Tumor susceptibility of Rassf1a knockout mice. Cancer Res 65(1):92–98. https://www.ncbi.nlm.nih.gov/pubmed/15665283. https://doi.org/10.1158/0008-5472.92.65.1
Ulitsky I, Bartel DP (2013) lincRNAs: Genomics, evolution, and mechanisms. Cell 154(1):26–46. https://doi.org/10.1016/j.cell.2013.06.020
Uno N, Hiramatsu K, Uno K, Komoto S, Kazuki Y, Oshimura M (2017) CRISPR/Cas9-induced transgene insertion and telomere-associated truncation of a single human chromosome for chromosome engineering in CHO and A9 cells. Sci Rep 7(1):12739. https://doi.org/10.1038/s41598-017-10418-7
Uno N, Abe S, Oshimura M, Kazuki Y (2018) Combinations of chromosome transfer and genome editing for the development of cell/animal models of human disease and humanized animal models. J Hum Genet 63(2):145–156. https://doi.org/10.1038/s10038-017-0378-7
van Steensel B, Furlong EEM (2019) The role of transcription in shaping the spatial organization of the genome. Nat Rev Mol Cell Biol 20(6):327–337. https://doi.org/10.1038/s41580-019-0114-6
Wang Y, Dan L, Li Q, Li L, Zhong L, Shao B, Yu F, He S, Tian S, He J, Xiao Q, Putti TC, He X, Feng Y, Lin Y, Xiang T (2019) ZMYND10, an epigenetically regulated tumor suppressor, exerts tumor-suppressive functions via miR145-5p/NEDD9 axis in breast cancer. Clin Epigenet 11(1):184. https://doi.org/10.1186/s13148-019-0785-z
Wang Y, Nie H, He X, Liao Z, Zhou Y, Zhou J, Ou C (2020) The emerging role of super enhancer-derived noncoding RNAs in human cancer. Theranostics 10(24):11049–11062. https://doi.org/10.7150/thno.49168
Zhang Q, He Y, Nie M, Cai W (2017) Roles of miR-138 and ISG15 in oral squamous cell carcinoma. Exp Ther Med 14(3):2329–2334. https://doi.org/10.3892/etm.2017.4720
Zheng H, Xie W (2019) The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 20(9):535–550. https://doi.org/10.1038/s41580-019-0132-4
Zhou LY, Zhang FW, Tong J, Liu F (2020) MiR-191–5p inhibits lung adenocarcinoma by repressing SATB1 to inhibit Wnt pathway. Mol Genet Genomic Med 8(1):e1043. https://doi.org/10.1002/mgg3.1043
Funding
This work was supported by the Japan Society for the Promotion of Science (JSPS) (Grant Number 19K10086 and 21K16213).
Author information
Authors and Affiliations
Contributions
T.O. and H. K. conceptualized the study, validated the data, performed formal analysis, and wrote the original draft. K. Y. provided the methodology and software and reviewed and edited the manuscript. H. K. performed project administration. H.K. acquired funding.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Responsible Editor: Tatsuo Fukagawa
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research was partly performed at the Tottori Bio Frontier Managed by Tottori Prefecture.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ohira, T., Yoshimura, K. & Kugoh, H. Human artificial chromosome carrying 3p21.3-p22.2 region suppresses hTERT transcription in oral cancer cells. Chromosome Res 31, 17 (2023). https://doi.org/10.1007/s10577-023-09726-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10577-023-09726-8