Abstract
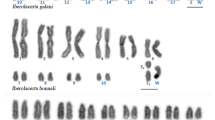
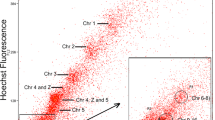
Geckos are a large group of lizards characterized by a rich variety of species, different modes of sex determination and diverse karyotypes. In spite of many unresolved questions on lizards’ phylogeny and taxonomy, the karyotypes of most geckos have been studied by conventional cytogenetic methods only. We used flow-sorted chromosome-specific painting probes of Japanese gecko (Gekko japonicus), Mediterranean house gecko (Hemidactylus turcicus) and flat-tailed house gecko (Hemidactylus platyurus) to reveal homologous regions and to study karyotype evolution in seven gecko species (Gekko gecko, G. japonicus, G. ulikovskii, G. vittatus, Hemidactylus frenatus, H. platyurus and H. turcicus). Generally, the karyotypes of geckos were found to be conserved, but we revealed some characteristic rearrangements including both fissions and fusions in Hemidactylus. The karyotype of H. platyurus contained a heteromorphic pair in all female individuals, where one of the homologues had a terminal DAPI-negative and C-positive heterochromatic block that might indicate a putative sex chromosome. Among two male individuals studied, only one carried such a polymorphism, and the second one had none, suggesting a possible ZZ/ZW sex determination in some populations of this species. We found that all Gekko species have retained the putative ancestral karyotype, whilst the fission of the largest ancestral chromosome occurred in the ancestor of modern Hemidactylus species. Three common fissions occurred in the ancestor of Mediterranean house and flat-tailed house geckos, suggesting their sister group relationships. PCR-assisted mapping on flow-sorted chromosome libraries with conserved DMRT1 gene primers in G. japonicus indicates the localization of DMRT1 gene on chromosome 6.







Similar content being viewed by others
Abbreviations
- 2n :
-
Diploid number of chromosomes
- ACA:
-
Anolis carolinensis
- DAPI:
-
4′,6-Diamidino-2-phenylindole
- DMRT1:
-
Doublesex- and mab-3-related transcription factor 1
- DOP-PCR:
-
Degenerate oligonucleotide primer–polymerase chain reaction
- FISH:
-
Fluorescence in situ hybridisation
- GJA:
-
Gekko japonicus
- GGE:
-
Gekko gecko
- GVI:
-
Gekko vittatus
- GUL:
-
Gekko ulikovskii
- HTU:
-
Hemidactylus turcicus
- HPL:
-
Hemidactylus platyurus
- HFR:
-
Hemidactylus frenatus
- NOR:
-
Nucleolar organizing region
References
Bansal R, Karanth PK (2010) Molecular phylogeny of Hemidactylus geckos (Squamata: Gekkonidae) of the Indian subcontinent reveals a unique Indian radiation and an Indian origin of Asian house geckos. Mol Phyl Evol 57:459–465
Brunner B, Hornung U, Shan Z et al (2001) Genomic organization and expression of the doublesex-related gene cluster in vertebrates and detection of putative regulatory regions for DMRT1. Genomics 77:8–17
Carranza S, Arnold EN (2006) Systematics, biogeography and evolution of Hemidactylus geckos (Reptilia: Gekkonidae) elucidated using mitochondrial DNA sequences. Mol Phyl Evol 38:531–545
Cohen MM, Huang CC, Clark HF (1967) The somatic chromosomes of 3 lizard species: Gekko gecko, Iguana iguana, and Crotaphytus collaris. Experimentia 15:769–771
De Smet WHO (1981) Description of the orcein stained kariotypes of 27 lizard species (Lacertilia: Reptilia) belonging to the familias Iguanidae, Agamidae, Chamaeleontidae and Gekkonidae (Ascalabota). Acta Zool Pathol Antverp 76:35–72
Ellegren H (2010) Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol 25:283–291
Ezaz T, Quinn AE, Miura I, Sarre D, Georges A, Graves JAM (2005) The dragon lizard Pogona vitticeps has ZZ/ZW micro-sex chromosomes. Chromosome Res 13:763–776
Ferguson-Smith MA, Trifonov V (2007) Mammalian karyotype evolution. Nat Rev Genet 8:950–962
Gamble T, Bauer AM, Greenbaum E, Jackman TR (2008a) Evidence for Gondwanan vicariance in an ancient clade of gecko lizards. J Biogeogr 35:88–104
Gamble T, Bauer AM, Greenbaum E, Jackman TR (2008b) Out of the blue: a novel, trans-Atlantic clade of gecko lizards (Gekkota, Squamata). Zool Scripta 37:355–366
Giovannotti M, Caputo V, O’Brien PCM, Lovell FL, Trifonov V, Cerioni PN, Olmo E, Ferguson-Smith MA, Rens W (2009) Skinks (Reptilia: Scincidae) have highly conserved karyotypes as revealed by chromosome painting. Cytogenet Genoime Res 127:224–231
Gorman GC (1973) The chromosomes of the Reptilia, a cytotaxonomic interpretation. In: Chiarelli AB, Capanna E (eds) Cytotaxonomy and vertebrate evolution. Academic, New York, pp 349–424
Han D, Zhou K, Bauer AM (2004) Phylogenetic relationships among the higher taxonomic categories of gekkotan lizards inferred from C-mos nuclear DNA sequences. Biol J Linn Soc 83:353–368
Howell WM, Black DA (1980) Controlled silver staining of nucleolus organizer regions with a protective colloidal developer: a 1 step method. Experientia 36:1014–1015
Kawai A, Ishijima J, Nishida C, Kosaka A, Ota H, Kohno S, Matsuda Y (2009) The ZW sex chromosomes of Gekko hokouensis (Gekkonidae, Squamata) represent highly conserved homology with those of avian species. Chromosoma 118:43–51
King M (1987) Monophyletism in the Gekkonidae: a chromosomal perspective. Aust J Zool 35:641–654
Kluge AG (1987) Cladistic relationships in the Gekkonoidea (Squamata, Sauria). Misc Publ Mus Zool univ Michigan 173(I–iv):1–54
Kumazawa Y (2007) Mitochondrial genomes from major lizard families suggest their phylogenetic relationships and ancient radiations. Gene 388:19–26
Kupriyanova LA, Darevsky IS, Ota H (1989) Karyotypic unidormity in the East Asian populations of Hemidactylus frenatus (Sauria: Gekkonidae). J Herpetol 23:294–296
Matsuda M, Nagahama Y, Shinomiya A, Sato T, Matsuda C, Kobayashi T, Morrey CE, Shibata N, Asakawa S, Shimizu N, Hori H, Hamaguchi S, Sakaizumi M (2002) DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417:559–563
Matsuda Y, Nishida-Umehara Ch, Tarui H et al (2005) Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosome Res 13:601–615
Matsuda Y, Nishida-Umehara CH, Tarui H, Kosaka A, Ishijima J, Matsubara K, Agata K (2007) Highly conserved linkage homology in the Archosauromorpha (turtles, crocodilians and birds) confirmed by comparative gene mapping. Chromosome Res 15:27
Moritz C (1984a) The evolution of a highly variable sex chromosome in Gehyra purpurascens (Gekkonidae). Chromosoma 90:111–119
Moritz C (1984b) The origin and evolution of parthenogenesis in Heteronotia binoei (Gekkonidae): chromosome banding studies. Chromosoma 89:151–162
Ogata M, Hasegawa Y, Ohtani H, Mineyama M, Miura I (2007) The ZZ/ZW sex-determining mechanism originated twice and independently during evolution of the frog. Rana rugosa Heredity 100:92–99
Olmo E (1986) A Reptilia. In: John B (ed) Animal cytogenetics 4, Chordata 3. Gebrueder Borntraeger, Berlin, pp 1–100
Olmo E, Signorino G (2005) Chromorep: a reptile chromosomes database. Internet references. http://ginux.univpm.it/scienze/chromorep/. Accessed 2 October 2005
Ota H, Matsui M, Hikida T, Hidaka T (1987) Karyotype of a gekkonid lizard, Cosymbotus platyurus, from Sabah, Borneo, Malaysia. Zool Sci 4:385–386
Ota H, Hikida T, Lue K (1989) Polyclony in a triploid gecko, Hemidactylus stejnegeri, from Taiwan, with notes on its bearing on the chromosomal diversity of the H. garnotii-vietnamensis complex (Sauria: Gekkonidae). Genetica 79:183–189
Pokorná M, Giovannotti M, Kratochvíl L, Kasai F, Trifonov VA, O’Brien PCM, Caputo V, Olmo E, Ferguson-Smith MA, Rens W (2011) Strong conservation of the bird Z chromosome in reptilian genomes is revealed by comparative painting despite 275 My divergence. Chromosoma 120:455–468
Rogatcheva MB, Serdyukova NA, Biltueva LS, Perelman PL, Borodin PM, Oda S, Graphodatsky AS (1997) Localization of the genes for major ribosomal RNA on chromosomes of the house musk shrew, Suncus murinus, at meiotic and mitotic cells by fluorescence in situ hybridization and silver staining. Genes Genet Syst 72:215–218
Schmid M, Felchtinger W, Nanda I, Schakowski R, Visbal Garcia R, Manzanilla Puppo J, Fernández Badillo A (1994) An extraordinarily low diploid chromosome number in the reptile Gonatodes taniae (Squamata, Gekkonidae). J Hered 85:255–260
Shetty S, Griffin DK, Graves JAM (1999) Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res 7:289–295
Shibaike Y, Takahashi Y, Arikura I et al (2009) Chromosome evolution in the lizard genus Gekko (Gekkonidae, Squamata, Reptilia) in the East Asian Islands. Cytogenet Genome Res 127:182–190
Smith HM (1935) Miscellaneous notes on Mexican lizards. Univ Kansas Sci Bull 22:119–155
Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie PG, Doran TJ, Sinclair AH (2009) The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461:267–271
Solleder E, Schmid M (1984) XX/XY-sex chromosomes in Gekko gecko (Sauria, Reptilia). Amphibia–Reptilia 5:339–345
Srikulnath K, Nishida C, Matsubara K, Uno Y, Thongpan A, Suputtitada S, Apisitwanich S, Matsuda Y (2009) Karyotypic evolution in squamate reptiles: comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res 17:975–986
Sumner A (1972) A simple method for demonstrating centromeric heterochromatin. Expl Cell Res 75:304–306
Telenius H, Pelmear AH, Tunnacliffe A, Carter NP, Behmel A, Ferguson-Smith MA, Nordenskjöld M, Pfragner R, Ponder BA (1992) Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosom Cancer 4:257–263
Tokunaga S (1985) Temperature-dependent sex determination in Gekko japonicus (Gekkonidae, Reptilia). Develop Growth Differ 27:117–120
Vidal N, Hedges SB (2005) The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C R Biologies 328:1000–1008
Vidal N, Hedges SB (2009) The molecular evolutionary tree of lizards, snakes, and amphisbaenians. C R Biologies 332:129–139
Volobouev V, Pasteur G, Ineich I, Dutrillaux B (1993) Chromosomal evidence for a hybrid origin of diploid parthenogenetic females from the unisexual–bisexual Lepidodactylus lugubris complex (Reptilia, Gekkonidae). Cytogenetics Cell Genet 63:194–199
Wang R, Ma K, Shi L, He W (1989) Studies on the mitotic karyotype and synoptonemal complex of Gekko gecko. Acta Metallurgica Sinica 10:271–275
Werner YL (1956) Chromosome numbers of some male geckos (Reptilia: Gekkonoidea). Bull Res Counc Israel 5B:319
Yang F, Carter NP, Shi L, Ferguson-Smith MA (1995) A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma 103:642–652, 31
Yang F, O’Brien P, Milne B, Graphodatsky A, Solansky N, Trifonov V, Rens W, Sargan D, Ferguson-Smith M (1999) A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 62:189–202
Yang F, Fu B, O’Brien PC, Robinson TJ, Ryder OA, Ferguson-Smith MA (2003) Karyotypic relationships of horses and zebras: results of cross-species chromosome painting. Cytogenet Genome Res 102:235–243
Yoshida MC, Itoh M (1974) Karyotype of the gekko Gekko japonicus. Chrom Info Serv 17:29–31
Yoshimoto S, Okada E, Umemoto H, Tamura K, Uno Y, Nishida-Umehara C, Matsuda Y, Takamatsu N, Shiba T, Ito M (2008) A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci USA 105:2469–2474
Acknowledgements
This work was funded by a Royal Society Grant to VT and MAF-S. VT was funded in part by MCB and SB RAS programmes. We wish to Fumio Kasai for establishing G. japonicus tissue culture. We would like to acknowledge anonymous reviewers for valuable comments.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Conly Rieder.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Conserved region of A. carolinensis DMRT1 first intron revealed by BLAST analysis of human DMRT1 sequence against Anolis sequence draft (AnoCar1.0) (a). The underlined parts were used for primer (DMRT1_ACA) design. b Sequence conservation of DMRT1_ACA region between chicken and green anole lizard, retrieved by BLAST analysis of Anolis DMRT1_ACA region against Gallus gallus on NCBI (JPEG 111 kb)
Supplementary Figure 2
Results of PCR screening of flow-sorted libraries of G. japonicus with DMRT1_ACA primers. Numbers indicate flow-sorted peak numbers. K Total genomic DNA of G. japonicus was used for PCR as a positive control. A strong band is seen on peak E, containing GJA6 (JPEG 143 kb)
Supplementary Table 1
Chromosome content of flow sorting peaks of H. platyurus, H. turcicus and G. japonicus (DOC 38 kb)
Rights and permissions
About this article
Cite this article
Trifonov, V.A., Giovannotti, M., O’Brien, P.C.M. et al. Chromosomal evolution in Gekkonidae. I. Chromosome painting between Gekko and Hemidactylus species reveals phylogenetic relationships within the group. Chromosome Res 19, 843–855 (2011). https://doi.org/10.1007/s10577-011-9241-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9241-4