Abstract
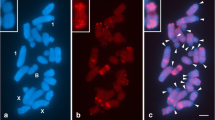
The quantitative variation of a conserved region of the LINE-1 ORF2 sequence was determined in eight species and subspecies of the subgenus Mus (M. m. domesticus, M. m. musculus, M. m. castaneus, M. spicilegus, M. spretus, M. cervicolor, M. cookii, M. caroli) and five Robertsonian races of M. m. domesticus. No differences in LINE-1 ORF2 content were found between all acrocentric or Robertsonian chromosome races, whereas the quantitative variation of the LINE-1 ORF2 sequences detected among the eight taxa partly matches with the clades into which the subgenus is divided. An accumulation of LINE-1 ORF2 elements likely occurred during the evolution of the subgenus. Within the Asiatic clade, M. cervicolor, cookii, and caroli show a low quantity of LINE-1 sequences, also detected within the Palearctic clade in M. m. castaneus and M. spretus, representing perhaps the ancestral condition within the subgenus. On the other hand, M. m. domesticus, M. m. musculus and M. spicilegus showed a high content of LINE-1 ORF2 sequences. Comparison between the chromosomal hybridization pattern of M. m. domesticus, which possesses the highest content, and M. spicilegus did not show any difference in the LINE-1 ORF2 distribution, suggesting that the quantitative variation of this sequence family did not involve chromosome restructuring or a preferential chromosome accumulation, during the evolution of M. m. domesticus.





Similar content being viewed by others
Abbreviations
- 3′UTR:
-
3′ untranslated region
- 5′UTR:
-
5′ untranslated region
- ANOVA:
-
analysis of variance
- bp:
-
base pair
- BSA:
-
bovine serum albumin
- DAPI:
-
4′,6-diamidino-2-phenylindole
- dCTP:
-
deoxycytidine triphosphate
- DNA:
-
deoxyribonucleic acid
- dNTP:
-
deoxyribonucleotide
- FISH:
-
fluorescence in-situ hybridization
- kb:
-
kilobase
- LINE-1:
-
long interspersed nuclear element-1
- LTR:
-
long terminal repeat
- MITE:
-
miniature inverted-repeat transposable element
- mtDNA:
-
mitochondrial deoxyribonucleic acid
- NIH:
-
National Institutes of Health
- ORF1:
-
open reading frame 1
- ORF2:
-
open reading frame 2
- PBS:
-
phosphate-buffered saline
- PCR:
-
polymerase chain reaction
- Rb:
-
Robertsonian
- RNA:
-
ribonucleic acid
- SDS:
-
sodium dodecyl sulfate
- SNPs:
-
single-nucleotide polymorphism
- SSC:
-
sodium chloride and sodium citrate
- TEs:
-
transposable elements
References
Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE (2008) Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res 18:869–880
Bailey JA, Carrel L, Chakravarti A, Eichler EE (2000) Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci U S A 97:6634–6639
Böhne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN (2008) Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res 16:203–215
Boissinot S, Davis J, Entezam A, Petrov D, Furano AV (2006) Fitness cost of LINE-1 (L1) activity in humans. Proc Natl Acad Sci U S A 103:9590–9594
Boyle AL, Ballard SG, Ward DC (1990) Differential distribution of long and short interspersed element sequences in the mouse genome: chromosome karyotyping by fluorescence in situ hybridization. Proc Natl Acad Sci U S A 87:7757–7761
Capanna E, Gropp A, Winking H, Noack G, Civitelli MV (1976) Robertsonian metacentrics in the mouse. Chromosoma 58:341–353
Castro JP, Carareto CM (2004) Drosophila melanogaster P transposable elements: mechanisms of transposition and regulation. Genetica 121:107–118
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Chevret P, Dobigny G (2005) Systematics and evolution of the subfamily Gerbillinae (Mammalia, Rodentia, Muridae). Mol Phylogenet Evol 35:674–688
Craig NL (1990) P element transposition. Cell 62:399–402
Deininger PL, Batzer MA (2002) Mammalian retroelements. Genome Res 12:1455–1465
Dobigny G, Ozouf-Costaz C, Waters PD, Bonillo C, Coutanceau JP, Volobouev V (2004) LINE-1 amplification accompanies explosive genome repatterning in rodents. Chromosome Res 12:787–793
Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH Jr (1991) Isolation of an active human transposable element. Science 254:1805–1808
Eide D, Anderson P (1988) Insertion and excision of Caenorhabditis elegans transposable element Tc1. Mol Cell Biol 8:737–746
Ferris SD, Sage RD, Prager EM, Ritte U, Wilson AC (1983) Mitochondrial DNA evolution in mice. Genetics 105:681–721
Feschotte C (2008) Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9:397–405
Finnegan DJ (1992) Transposable elements. Curr Opin Genet Dev 2:861–867
Furano AV (2000) The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Prog Nucleic Acid Res Mol Biol 64:255–294
Furano AV, Duvernell DD, Boissinot S (2004) L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet 20:9–14
Garagna S, Redi CA, Capanna E et al (1993) Genome distribution, chromosomal allocation, and organization of the major and minor satellite DNAs in 11 species and subspecies of the genus Mus. Cytogenet Cell Genet 64:247–255
Gray YH (2000) It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet 16:461–468
Gropp A, Winking H (1981) Robertsonian translocations: cytology, meiosis segregation pattern and biological consequences of heterozygosity. In: Berry RJ (ed) Biology of the house mouse. Academic Press, pp 141–170
Guénet JL, Bonhomme F (2003) Wild mice: an ever-increasing contribution to a popular mammalian model. Trends Genet 19:24–31
Hauffe HC, Searle JB (1998) Chromosomal heterozygosity and fertility in house mice (Mus musculus domesticus) from Northern Italy. Genetics 150:1143–1154
Kazazian HH Jr (1998) Mobile elements and disease. Curr Opin Genet Dev 8:343–350
Kazazian HH Jr (1999) An estimated frequency of endogenous insertional mutations in humans. Nat Genet 22:130
Kazazian HH Jr (2000) Genetics. L1 retrotransposons shape the mammalian genome. Science 289:1152–1153
Kehrer-Sawatzki H, Schreiner B, Tänzer S, Platzer M, Müller S, Hameister H (2002) Molecular characterization of the pericentric inversion that causes differences between chimpanzee chromosome 19 and human chromosome 17. Am J Hum Genet 71:375–388
Kidwell MG (2002) Transposable elements and the evolution of genome size in eukaryotes. Genetica 115:49–63
Kidwell MG, Lisch D (1997) Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci U S A 94:7704–7711
Lander ES, Linton LM, Birren B et al (2001) International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409:860–921
Loeb DD, Padgett RW, Hardies SC et al (1986) The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol 6:168–182
Lundrigan BL, Jansa SA, Tucker PK (2002) Phylogenetic relationships in the genus mus, based on paternally, maternally, and biparentally inherited characters. Syst Biol 51:410–431
Lyon MF (2000) LINE-1 elements and X chromosome inactivation: a function for “junk” DNA? Proc Natl Acad Sci U S A 97:6248–6249
Marchal JA, Acosta MJ, Bullejos M, Puerma E, Díaz de la Guardia R, Sánchez A (2006) Distribution of L1-retroposons on the giant sex chromosomes of Microtus cabrerae (Arvicolidae, Rodentia): functional and evolutionary implications. Chromosome Res 14:177–186
Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A (1991) Reverse transcriptase encoded by a human transposable element. Science 254:1808–1810
Muotri AR, Marchetto MC, Coufal NG, Gage FH (2007) The necessary junk: new functions for transposable elements. Hum Mol Genet 16(Spec No. 2):R159–R167
Prager EM, Tichy H, Sage RD (1996) Mitochondrial DNA sequence variation in the eastern house mouse, Mus musculus: Comparison with other house mice and report of a 75-bp tandem repeat. Genetics 143:427–446
Rebuzzini P, Martinelli P, Blasco M, Giulotto E, Mondello C (2007) Inhibition of gene amplification in telomerase deficient immortalized mouse embryonic fibroblasts. Carcinogenesis 28:553–559
Redi CA, Garagna S, Della Valle G (1990) Differences in the organization and chromosomal allocation of satellite DNA between the European long tailed house mice Mus domesticus and Mus musculus. Chromosoma 99:11–17
Sassaman DM, Dombroski BA, Moran JV et al (1997) Many human L1 elements are capable of retrotransposition. Nat Genet 16:37–43
Schwartz A, Chan DC, Brown LG et al (1998) Reconstructing hominid Y evolution: X-homologous block, created by X-Y transposition, was disrupted by Yp inversion through LINE-LINE recombination. Hum Mol Genet 7:1–11
She JX, Bonhomme F, Boursot P, Thaler L, Catzeflis F (1990) Molecular phylogenies in the Genus Mus—comparative analysis of electrophoretic, ScnDNA hybridization, and MtDNA RFLP data. Biol J Linn Soc 41:83–103
Solano E, Castiglia R, Corti M (2007) A new chromosomal race of the house mouse—Mus musculus domesticus—in the Vulcano Island-Aeolian Archipelago, Italy. Hereditas 144:75–77
Song M, Boissinot S (2007) Selection against LINE-1 retrotransposons results principally from their ability to mediate ectopic recombination. Gene 390:206–213
Spradling AC (1994) Transposable elements and the evolution of heterochromatin. Soc Gen Physiol Ser 49:69–83
Tucker PK, Sandstedt SA, Lundrigan BL (2005) Phylogenetic relationships in the subgenus Mus (genus Mus, family Muridae, subfamily Murinae): examining gene trees and species trees. Biol J Linn Soc 84:653–662
Usdin K, Chevret P, Catzeflis FM, Verona R, Furano AV (1995) L1 (LINE-1) retrotransposable elements provide a “fossil” record of the phylogenetic history of murid rodents. Mol Biol Evol 12:73–82
Vos JC, van Luenen HG, Plasterk RH (1993) Characterization of the Caenorhabditis elegans Tc1 transposase in vivo and in vitro. Genes Dev 7:1244–1253
Waters PD, Dobigny G, Pardini AT, Robinson TJ (2004) LINE-1 distribution in Afrotheria and Xenarthra: implications for understanding the evolution of LINE-1 in eutherian genomes. Chromosoma 113:137–144
Waterston RH, Lindblad-Toh K, Birney E et al (2002) Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520–562
Weiner AM (2002) SINEs and LINEs: the art of biting the hand that feeds you. Curr Opin Cell Biol 14:343–350
Wessler SR, Bureau TE, White SE (1995) LTR-retrotransposons and MITEs: important players in the evolution of plant genomes. Curr Opin Genet Dev 5:814–821
Yonekawa H, Moriwaki K, Gotoh O et al (1982) Origins of laboratory mice deduced from restriction patterns of mitochondrial DNA. Differentiation 22:222–226
Acknowledgements
This work was supported by PRIN-COFIN 2005, Cariplo Foundation and FIRB 2005 (Project N.RBIP06FH7J) (Italy).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Walther Traut.
Rights and permissions
About this article
Cite this article
Rebuzzini, P., Castiglia, R., Nergadze, S.G. et al. Quantitative variation of LINE-1 sequences in five species and three subspecies of the subgenus Mus and in five Robertsonian races of Mus musculus domesticus . Chromosome Res 17, 65–76 (2009). https://doi.org/10.1007/s10577-008-9004-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-008-9004-z