Abstract
Purpose
Several cancer subtypes (pancreatic, breast, liver, and colorectal) rapidly advance to higher aggressive stages in diabetes. Though hyperglycemia has been considered as a fuel for growth of cancer cells, pathways leading to this condition are still under investigation. Cellular polyamines can modulate normal and cancer cell growth, and inhibitors of polyamine synthesis have been approved for treating colon cancer, however the role of polyamines in diabetes-mediated cancer advancement is unclear as yet. We hypothesized that polyamine metabolic pathway is involved with increased proliferation of breast cancer cells under high glucose (HG) conditions.
Methods
Studies were performed with varying concentrations of glucose (5–25 mM) exposure in invasive, triple negative breast cancer cells, MDA-MB-231; non-invasive, estrogen/progesterone receptor positive breast cancer cells, MCF-7; and non-tumorigenic mammary epithelial cells, MCF-10A.
Results
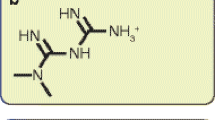
There was a significant increase in proliferation with HG (25 mM) at 48–72 h in both MDA-MB-231 and MCF-10A cells but no such effect was observed in MCF-7 cells. This was correlated to higher activity of ornithine decarboxylase (ODC), a rate-limiting enzyme in polyamine synthesis pathway. Inhibitor of polyamine synthesis (difluoromethylornithine, DFMO, 5 mM) was quite effective in suppressing HG-mediated cell proliferation and ODC activity in MDA-MB-231 and MCF-10A cells. Polyamine (putrescine) levels were significantly elevated with HG treatment in MDA-MB-231 cells. HG exposure also increased the metastasis of MDA-MB-231 cells.
Conclusions
Our cellular findings indicate that polyamine inhibition should be explored in patient population as a target for future chemotherapeutics in diabetic breast cancer.








Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Onitilo AA, Engel JM, Glurich I, Stankowski RV, Williams GM, Doi SA (2012) Diabetes and cancer I: risk, survival, and implications for screening. Cancer Causes Control 23(6):967–981
Onitilo AA, Stankowski RV, Berg RL, Engel JM, Glurich I, Williams GM, Doi SA (2014) Breast cancer incidence before and after diagnosis of type 2 diabetes mellitus in women: increased risk in the prediabetes phase. Eur J Cancer Prev 23(2):76–83
Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, Pollak M, Regensteiner JG, Yee D (2010) Diabetes and cancer: a consensus report. CA Cancer J Clin 60(4):207–221
Chowdhury TA (2010) Diabetes and cancer. QJM 103(12):905–915
Xu CX, Zhu HH, Zhu YM (2014) Diabetes and cancer: associations, mechanisms, and implications for medical practice. World J Diabetes 5(3):372–380
Ryu TY, Park J, Scherer PE (2014) Hyperglycemia as a risk factor for cancer progression. Diabetes Metab J 38(5):330–336
Canizares F, Salinas J, de las Heras M, Diaz J, Tovar I, Martinez P, Penafiel R (1999) Prognostic value of ornithine decarboxylase and polyamines in human breast cancer: correlation with clinicopathologic parameters. Clin Cancer Res 5(8):2035–2041
Celik VK, Kapancik S, Kacan T, Kacan SB, Kapancik S, Kilicgun H (2017) Serum levels of polyamine synthesis enzymes increase in diabetic patients with breast cancer. Endocr Connect 6(8):574–579
Geck RC, Foley JR, Murray Stewart T, Asara JM, Casero RA Jr, Toker A (2020) Inhibition of the polyamine synthesis enzyme ornithine decarboxylase sensitizes triple-negative breast cancer cells to cytotoxic chemotherapy. J Biol Chem 295(19):6263–6277
Igarashi K, Kashiwagi K (2010) Modulation of cellular function by polyamines. Int J Biochem Cell Biol 42(1):39–51
Paz EA, Garcia-Huidobro J, Ignatenko NA (2011) Polyamines in cancer. Adv Clin Chem 54:45–70
Casero RA Jr, Murray Stewart T, Pegg AE (2018) Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat Rev Cancer 18(11):681–695
Thomas T, Thomas TJ (2001) Polyamines in cell growth and cell death: molecular mechanisms and therapeutic applications. Cell Mol Life Sci 58(2):244–258
Auvinen M, Paasinen A, Andersson LC, Holtta E (1992) Ornithine decarboxylase activity is critical for cell transformation. Nature 360(6402):355–358
Bartholeyns J (1983) Treatment of metastatic Lewis lung carcinoma with DL-alpha-difluoromethylornithine. Eur J Cancer Clin Oncol 19(4):567–572
Gerner EW, Meyskens FL Jr. (2004) Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer 4(10):781–792
Nakanishi S, Cleveland JL (2016) Targeting the polyamine-hypusine circuit for the prevention and treatment of cancer. Amino Acids 48(10):2353–2362
Pegg AE (1988) Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy. Cancer Res 48(4):759–774
Shantz LM, Levin VA (2007) Regulation of ornithine decarboxylase during oncogenic transformation: mechanisms and therapeutic potential. Amino Acids 33(2):213–223
Gilmour SK (2007) Polyamines and nonmelanoma skin cancer. Toxicol Appl Pharmacol 224(3):249–256
Kingsnorth AN, Wallace HM, Bundred NJ, Dixon JM (1984) Polyamines in breast cancer. Br J Surg 71(5):352–356
Milovic V, Turchanowa L (2003) Polyamines and colon cancer. Biochem Soc Trans 31(2):381–383
Rhodes DR, Barrette TR, Rubin MA, Ghosh D, Chinnaiyan AM (2002) Meta-analysis of microarrays: interstudy validation of gene expression profiles reveals pathway dysregulation in prostate cancer. Cancer Res 62(15):4427–4433
Park MH, Igarashi K (2013) Polyamines and their metabolites as diagnostic markers of human diseases. Biomol Ther (Seoul) 21(1):1–9
Casero RA Jr, Marton LJ (2007) Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov 6(5):373–390
Soda K (2011) The mechanisms by which polyamines accelerate tumor spread. J Exp Clin Cancer Res 30:95
Ma H, Li Q, Wang J, Pan J, Su Z, Lu S (2021) Dual inhibition of ornithine decarboxylase and A 1 adenosine receptor efficiently suppresses breast tumor cells. Front Oncol. https://doi.org/10.3389/fonc.2021.636373
Sweeney C (2020) Targeting the polyamine pathway—“a means” to overcome chemoresistance in triple-negative breast cancer. J Biol Chem 295(19):6278–6279
Lee J, Sperandio V, Frantz DE, Longgood J, Camilli A, Phillips MA, Michael AJ (2009) An alternative polyamine biosynthetic pathway is widespread in bacteria and essential for biofilm formation in Vibrio cholerae. J Biol Chem 284(15):9899–9907
Liu L, Guo X, Rao JN, Zou T, Marasa BS, Chen J, Greenspon J, Casero RA Jr., Wang JY (2006) Polyamine-modulated c-Myc expression in normal intestinal epithelial cells regulates p21Cip1 transcription through a proximal promoter region. Biochem J 398(2):257–267
Gupta C, Tikoo K (2013) High glucose and insulin differentially modulates proliferation in MCF-7 and MDA-MB-231 cells. J Mol Endocrinol 51(1):119–129
Lopez R, Arumugam A, Joseph R, Monga K, Boopalan T, Agullo P, Gutierrez C, Nandy S, Subramani R, de la Rosa JM, Lakshmanaswamy R (2013) Hyperglycemia enhances the proliferation of non-tumorigenic and malignant mammary epithelial cells through increased leptin/IGF1R signaling and activation of AKT/mTOR. PLoS ONE 8(11):e79708
Masur K, Vetter C, Hinz A, Tomas N, Henrich H, Niggemann B, Zanker KS (2011) Diabetogenic glucose and insulin concentrations modulate transcriptome and protein levels involved in tumour cell migration, adhesion and proliferation. Br J Cancer 104(2):345–352
Wairagu PM, Phan AN, Kim MK, Han J, Kim HW, Choi JW, Kim KW, Cha SK, Park KH, Jeong Y (2015) Insulin priming effect on estradiol-induced breast cancer metabolism and growth. Cancer Biol Ther 16(3):484–492
Flores-Lopez LA, Martinez-Hernandez MG, Viedma-Rodriguez R, Diaz-Flores M, Baiza-Gutman LA (2016) High glucose and insulin enhance uPA expression, ROS formation and invasiveness in breast cancer-derived cells. Cell Oncol (Dordr) 39(4):365–378
Ruiz-Perez MV, Medina MA, Urdiales JL, Keinanen TA, Sanchez-Jimenez F (2015) Polyamine metabolism is sensitive to glycolysis inhibition in human neuroblastoma cells. J Biol Chem 290(10):6106–6119
Acknowledgements
Research reported in this publication was supported by NIH Grants: GM103427 (SC), CA204345 (RAC), and CA235863 (RAC). In addition, funding was received from Nebraska Research Initiative Collaborative Seed Grant (SC), and Samuel Waxman Cancer Research Foundation (RAC). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Cell culture facility and fluorescence microscopy facilities at UNK were supported by grants from the National Institute for General Medical Science (NIGMS) (5P20GM103427), a component of the National Institutes of Health (NIH), as well as Nebraska Research Initiative. HT was supported as graduate research assistant through financial support provided by the Division of Research and the Office of Graduate Studies and Academic Outreach at University of Nebraska at Kearney.
Author information
Authors and Affiliations
Contributions
CC, the primary author was involved with project design, experimentation, and writing the manuscript; JO and JM performed cellular assays; MD under the supervision of RAC performed enzyme activity assays at John Hopkins; HT edited the manuscript; and SC and RAC supervised the work, helped with project design, and edited the manuscript. We thank Dr Sophie Alvarez at the Proteomics & Metabolomics Facility, Nebraska Center for Biotechnology at the University of Nebraska-Lincoln for the polyamines analysis. The facility and instrumentation are supported by the Nebraska Research Initiative.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest to disclose.
Consent for publication
This manuscript is being submitted upon consent from all authors listed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Capellen, C.C., Ortega-Rodas, J., Morwitzer, M.J. et al. Hyperglycemic conditions proliferate triple negative breast cancer cells: role of ornithine decarboxylase. Breast Cancer Res Treat 190, 255–264 (2021). https://doi.org/10.1007/s10549-021-06388-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-021-06388-0