Abstract
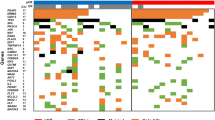
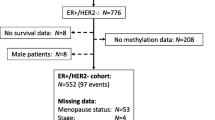
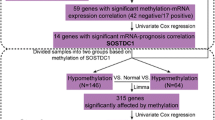
Identification of prognostic gene expression signatures may enable improved decisions about management of breast cancer. To identify a prognostic signature for breast cancer, we performed DNA methylation profiling and identified methylation markers that were associated with expression of ER, PR, HER2, CK5/6, and EGFR proteins. Methylation markers that were correlated with corresponding mRNA expression levels were identified using 208 invasive tumors from a population-based case–control study conducted in Poland. Using this approach, we defined the methylation expression index (MEI) signature that was based on a weighted sum of mRNA levels of 57 genes. Classification of cases as low or high MEI scores was related to survival using Cox regression models. In the Polish study, women with ER-positive low MEI cancers had reduced survival at a median of 5.20 years of follow-up, HR = 2.85 95 % CI = 1.25–6.47. Low MEI was also related to decreased survival in four independent datasets totaling over 2500 ER-positive breast cancers. These results suggest that integrated analysis of tumor expression markers, DNA methylation, and mRNA data may be an important approach for identifying breast cancer prognostic signatures. Prospective assessment of MEI along with other prognostic signatures should be evaluated in future studies.



Similar content being viewed by others
References
Laird PW, Jaenisch R (1994) DNA methylation and cancer. Hum Mol Genet 3:1487–1495
Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, Davies SR, Snider J, Stijleman IJ, Reed J, Cheang MC, Mardis ER, Perou CM, Bernard PS, Ellis MJ (2010) A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res 16(21):5222–5232. doi:10.1158/1078-0432.CCR-10-1282
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351(27):2817–2826. doi:10.1056/NEJMoa041588
van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415(6871):530–536. doi:10.1038/415530a
Veeck J, Esteller M (2010) Breast cancer epigenetics: from DNA methylation to microRNAs. J Mammary Gland Biol Neoplasia 15(1):5–17. doi:10.1007/s10911-010-9165-1
Widschwendter M, Jones PA (2002) DNA methylation and breast carcinogenesis. Oncogene 21(35):5462–5482. doi:10.1038/sj.onc.1205606
Bediaga NG, Acha-Sagredo A, Guerra I, Viguri A, Albaina C, Ruiz Diaz I, Rezola R, Alberdi MJ, Dopazo J, Montaner D, de Renobales M, Fernandez AF, Field JK, Fraga MF, Liloglou T, de Pancorbo MM (2010) DNA methylation epigenotypes in breast cancer molecular subtypes. Breast Cancer Res 12(5):R77. doi:10.1186/bcr2721
Christensen BC, Kelsey KT, Zheng S, Houseman EA, Marsit CJ, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Kushi LH, Kwan ML, Wiencke JK (2010) Breast cancer DNA methylation profiles are associated with tumor size and alcohol and folate intake. PLoS Genet 6(7):e1001043. doi:10.1371/journal.pgen.1001043
Dedeurwaerder S, Desmedt C, Calonne E (2011) Largest ever DNA methylation dataset for breast cancer completed. Expert Rev Mol Diagn 11(5):470
Fackler MJ, Umbricht CB, Williams D, Argani P, Cruz LA, Merino VF, Teo WW, Zhang Z, Huang P, Visvananthan K, Marks J, Ethier S, Gray JW, Wolff AC, Cope LM, Sukumar S (2011) Genome-wide methylation analysis identifies genes specific to breast cancer hormone receptor status and risk of recurrence. Cancer Res 71(19):6195–6207. doi:10.1158/0008-5472.CAN-11-1630
Fang F, Turcan S, Rimner A, Kaufman A, Giri D, Morris LG, Shen R, Seshan V, Mo Q, Heguy A, Baylin SB, Ahuja N, Viale A, Massague J, Norton L, Vahdat LT, Moynahan ME, Chan TA (2011) Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med 3(75):75ra25. doi:10.1126/scitranslmed.3001875
Flanagan JM, Cocciardi S, Waddell N, Johnstone CN, Marsh A, Henderson S, Simpson P, da Silva L, Khanna K, Lakhani S, Boshoff C, Chenevix-Trench G (2010) DNA methylome of familial breast cancer identifies distinct profiles defined by mutation status. Am J Hum Genet 86(3):420–433. doi:10.1016/j.ajhg.2010.02.008
Hill VK, Ricketts C, Bieche I, Vacher S, Gentle D, Lewis C, Maher ER, Latif F (2011) Genome-wide DNA methylation profiling of CpG islands in breast cancer identifies novel genes associated with tumorigenicity. Cancer Res 71(8):2988–2999. doi:10.1158/0008-5472.CAN-10-4026
Holm K, Hegardt C, Staaf J, Vallon-Christersson J, Jonsson G, Olsson H, Borg A, Ringner M (2010) Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast Cancer Res 12(3):R36. doi:10.1186/bcr2590
Kamalakaran S, Varadan V, Giercksky Russnes HE, Levy D, Kendall J, Janevski A, Riggs M, Banerjee N, Synnestvedt M, Schlichting E, Karesen R, Shama Prasada K, Rotti H, Rao R, Rao L, Eric Tang MH, Satyamoorthy K, Lucito R, Wigler M, Dimitrova N, Naume B, Borresen-Dale AL, Hicks JB (2011) DNA methylation patterns in luminal breast cancers differ from non-luminal subtypes and can identify relapse risk independent of other clinical variables. Mol Oncol 5(1):77–92. doi:10.1016/j.molonc.2010.11.002
Li L, Lee KM, Han W, Choi JY, Lee JY, Kang GH, Park SK, Noh DY, Yoo KY, Kang D (2010) Estrogen and progesterone receptor status affect genome-wide DNA methylation profile in breast cancer. Hum Mol Genet 19(21):4273–4277. doi:10.1093/hmg/ddq351
Ronneberg JA, Fleischer T, Solvang HK, Nordgard SH, Edvardsen H, Potapenko I, Nebdal D, Daviaud C, Gut I, Bukholm I, Naume B, Borresen-Dale AL, Tost J, Kristensen V (2011) Methylation profiling with a panel of cancer related genes: association with estrogen receptor, TP53 mutation status and expression subtypes in sporadic breast cancer. Mol Oncol 5(1):61–76. doi:10.1016/j.molonc.2010.11.004
Van der Auwera I, Yu W, Suo L, Van Neste L, van Dam P, Van Marck EA, Pauwels P, Vermeulen PB, Dirix LY, Van Laere SJ (2010) Array-based DNA methylation profiling for breast cancer subtype discrimination. PLoS ONE 5(9):e12616. doi:10.1371/journal.pone.0012616
Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70. doi:10.1038/nature11412
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486(7403):346–352. doi:10.1038/nature10983
Kristensen VN, Lingjaerde OC, Russnes HG, Vollan HK, Frigessi A, Borresen-Dale AL (2014) Principles and methods of integrative genomic analyses in cancer. Nat Rev Cancer 14(5):299–313. doi:10.1038/nrc3721
Yang XR, Sherman ME, Rimm DL, Lissowska J, Brinton LA, Peplonska B, Hewitt SM, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Cartun R, Mandich D, Rymkiewicz G, Ligaj M, Lukaszek S, Kordek R, Garcia-Closas M (2007) Differences in risk factors for breast cancer molecular subtypes in a population-based study. Cancer Epidemiol Biomarkers Prev 16(3):439–443. doi:10.1158/1055-9965.EPI-06-0806
Carey LA, Perou CM, Livasy CA, Dressler LG, Cowan D, Conway K, Karaca G, Troester MA, Tse CK, Edmiston S, Deming SL, Geradts J, Cheang MC, Nielsen TO, Moorman PG, Earp HS, Millikan RC (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295(21):2492–2502. doi:10.1001/jama.295.21.2492
Garcia-Closas M, Brinton LA, Lissowska J, Chatterjee N, Peplonska B, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Blair A, Kalaylioglu Z, Rymkiewicz G, Mazepa-Sikora D, Kordek R, Lukaszek S, Sherman ME (2006) Established breast cancer risk factors by clinically important tumour characteristics. Br J Cancer 95(1):123–129. doi:10.1038/sj.bjc.6603207
Sherman ME, Rimm DL, Yang XR, Chatterjee N, Brinton LA, Lissowska J, Peplonska B, Szeszenia-Dabrowska N, Zatonski W, Cartun R, Mandich D, Rymkiewicz G, Ligaj M, Lukaszek S, Kordek R, Kalaylioglu Z, Harigopal M, Charrette L, Falk RT, Richesson D, Anderson WF, Hewitt SM, Garcia-Closas M (2007) Variation in breast cancer hormone receptor and HER2 levels by etiologic factors: a population-based analysis. Int J Cancer 121(5):1079–1085. doi:10.1002/ijc.22812
Kaplan EL, Meier P (1958) Nonparametric estimation from incomplete observations. J Am Stat Assoc 53:457–481
Cox DR (1972) Regression models and life-tables (with discussion). J R Stat Soc Ser C Appl Stat Series B 34:187–220
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347(25):1999–2009. doi:10.1056/NEJMoa021967
Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, Klijn JG, Larsimont D, Buyse M, Bontempi G, Delorenzi M, Piccart MJ, Sotiriou C (2007) Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol 25(10):1239–1246. doi:10.1200/JCO.2006.07.1522
Killian JK, Bilke S, Davis S, Walker RL, Jaeger E, Killian MS, Waterfall JJ, Bibikova M, Fan JB, Smith WI Jr, Meltzer PS (2011) A methyl-deviator epigenotype of estrogen receptor-positive breast carcinoma is associated with malignant biology. Am J Pathol 179(1):55–65. doi:10.1016/j.ajpath.2011.03.022
Gianni L, Zambetti M, Clark K, Baker J, Cronin M, Wu J, Mariani G, Rodriguez J, Carcangiu M, Watson D, Valagussa P, Rouzier R, Symmans WF, Ross JS, Hortobagyi GN, Pusztai L, Shak S (2005) Gene expression profiles in paraffin-embedded core biopsy tissue predict response to chemotherapy in women with locally advanced breast cancer. J Clin Oncol 23(29):7265–7277. doi:10.1200/JCO.2005.02.0818
Yan PS, Venkataramu C, Ibrahim A, Liu JC, Shen RZ, Diaz NM, Centeno B, Weber F, Leu YW, Shapiro CL, Eng C, Yeatman TJ, Huang TH (2006) Mapping geographic zones of cancer risk with epigenetic biomarkers in normal breast tissue. Clin Cancer Res 12(22):6626–6636. doi:10.1158/1078-0432.CCR-06-0467
Sandhu R, Rivenbark AG, Mackler RM, Livasy CA, Coleman WB (2014) Dysregulation of microRNA expression drives aberrant DNA hypermethylation in basal-like breast cancer. Int J Oncol 44(2):563–572. doi:10.3892/ijo.2013.2197
Roll JD, Rivenbark AG, Sandhu R, Parker JS, Jones WD, Carey LA, Livasy CA, Coleman WB (2013) Dysregulation of the epigenome in triple-negative breast cancers: basal-like and claudin-low breast cancers express aberrant DNA hypermethylation. Exp Mol Pathol 95(3):276–287. doi:10.1016/j.yexmp.2013.09.001
Stefansson OA, Jonasson JG, Olafsdottir K, Hilmarsdottir H, Olafsdottir G, Esteller M, Johannsson OT, Eyfjord JE (2011) CpG island hypermethylation of BRCA1 and loss of pRb as co-occurring events in basal/triple-negative breast cancer. Epigenetics 6(5):638–649. doi:10.4161/epi.6.5.15667
Matuschek C, Bolke E, Lammering G, Gerber PA, Peiper M, Budach W, Taskin H, Prisack HB, Schieren G, Orth K, Bojar H (2010) Methylated APC and GSTP1 genes in serum DNA correlate with the presence of circulating blood tumor cells and are associated with a more aggressive and advanced breast cancer disease. Eur J Med Res 15:277–286
Yamada N, Nishida Y, Tsutsumida H, Hamada T, Goto M, Higashi M, Nomoto M, Yonezawa S (2008) MUC1 expression is regulated by DNA methylation and histone H3 lysine 9 modification in cancer cells. Cancer Res 68(8):2708–2716. doi:10.1158/0008-5472.CAN-07-6844
Zrihan-Licht S, Weiss M, Keydar I, Wreschner DH (1995) DNA methylation status of the MUC1 gene coding for a breast-cancer-associated protein. Int J Cancer 62(3):245–251
Yu Z, Wang L, Wang C, Ju X, Wang M, Chen K, Loro E, Li Z, Zhang Y, Wu K, Casimiro MC, Gormley M, Ertel A, Fortina P, Chen Y, Tozeren A, Liu Z, Pestell RG (2013) Cyclin D1 induction of Dicer governs microRNA processing and expression in breast cancer. Nat Commun 4:2812. doi:10.1038/ncomms3812
Cowling VH (2010) Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene 29(6):930–936. doi:10.1038/onc.2009.368
Liu T, Niu Y, Feng Y, Niu R, Yu Y, Lv A, Yang Y (2008) Methylation of CpG islands of p16(INK4a) and cyclinD1 overexpression associated with progression of intraductal proliferative lesions of the breast. Hum Pathol 39(11):1637–1646. doi:10.1016/j.humpath.2008.04.001
Sizemore ST, Sizemore GM, Booth CN, Thompson CL, Silverman P, Bebek G, Abdul-Karim FW, Avril S, Keri RA (2014) Hypomethylation of the MMP7 promoter and increased expression of MMP7 distinguishes the basal-like breast cancer subtype from other triple-negative tumors. Breast Cancer Res Treat 146(1):25–40. doi:10.1007/s10549-014-2989-4
Chen L, Zhou XG, Zhou XY, Zhu C, Ji CB, Shi CM, Qiu J, Guo XR (2013) Overexpression of C10orf116 promotes proliferation, inhibits apoptosis and enhances glucose transport in 3T3-L1 adipocytes. Mol Med Rep 7(5):1477–1481. doi:10.3892/mmr.2013.1351
Simpson NE, Lambert WM, Watkins R, Giashuddin S, Huang SJ, Oxelmark E, Arju R, Hochman T, Goldberg JD, Schneider RJ, Reiz LF, Soares FA, Logan SK, Garabedian MJ (2010) High levels of Hsp90 cochaperone p23 promote tumor progression and poor prognosis in breast cancer by increasing lymph node metastases and drug resistance. Cancer Res 70(21):8446–8456. doi:10.1158/0008-5472.CAN-10-1590
Author information
Authors and Affiliations
Corresponding author
Additional information
Jonine D. Figueroa and Howard Yang have equally contributed to this work.
Mark E. Sherman and Maxwell Lee are Co-senior authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Figueroa, J.D., Yang, H., Garcia-Closas, M. et al. Integrated analysis of DNA methylation, immunohistochemistry and mRNA expression, data identifies a methylation expression index (MEI) robustly associated with survival of ER-positive breast cancer patients. Breast Cancer Res Treat 150, 457–466 (2015). https://doi.org/10.1007/s10549-015-3314-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-015-3314-6