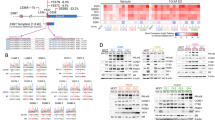
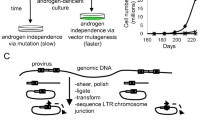
Abstract
Endocrine treatment of breast cancer is widely applied and effective. However, in advanced disease cases, the tumors will eventually progress into an estrogen-independent and therapy-resistant phenotype. To elucidate the molecular mechanisms underlying this endocrine therapy failure, we applied retroviral insertion mutagenesis to identify the main genes conferring estrogen independence to human breast cancer cells. Estrogen-dependent ZR-75-1 cells were infected with replication-defective retroviruses followed by selection with the anti-estrogen 4-hydroxy-tamoxifen. In the resulting panel of 79 tamoxifen-resistant cell lines, the viral integrations were mapped within the human genome. Genes located in the immediate proximity of the retroviral integration sites were characterized for altered expression and their capacity to confer anti-estrogen resistance when transfected into breast cancer cells. Out of 15 candidate BCAR (breast cancer anti-estrogen resistance) genes, seven (AKT1, AKT2, BCAR1, BCAR3, EGFR, GRB7, and TRERF1/BCAR2) were shown to directly underlie estrogen independence. Our results show that insertion mutagenesis is a powerful tool to identify BCAR loci, which may provide insights into the molecular and cellular mechanisms of breast tumor progression and therapy resistance thereby offering novel targets for the development of tailor-made therapeutical and prevention strategies.


Similar content being viewed by others
References
Early Breast Cancer Trialists’ Collaborative Group (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365:1687–1717
Jaiyesimi IA, Buzdar AU, Decker DA et al (1995) Use of tamoxifen for breast cancer: twenty-eight years later. J Clin Oncol 13:513–529
Ali S, Coombes RC (2002) Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer 2:101–112
Clarke R, Liu MC, Bouker KB et al (2003) Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene 22:7316–7339
Osborne CK, Shou J, Massarweh S et al (2005) Crosstalk between estrogen receptor and growth factor receptor pathways as a cause for endocrine therapy resistance in breast cancer. Clin Cancer Res 11:865s–870s
Lewis JS, Jordan VC (2005) Selective estrogen receptor modulators (SERMs): mechanisms of anticarcinogenesis and drug resistance. Mutat Res 591:247–263
Riggins RB, Schrecengost RS, Guerrero MS et al (2007) Pathways to tamoxifen resistance. Cancer Lett doi:10.106/j.canlet.2007.2003.2016
van de Vijver MJ, He YD, van’t Veer LJ et al (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347:1999–2009
Paik S, Shak S, Tang G et al (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826
Jansen MP, Foekens JA, van Staveren IL et al (2005) Molecular classification of tamoxifen-resistant breast carcinomas by gene expression profiling. J Clin Oncol 23:732–740
Foekens JA, Atkins D, Zhang Y et al (2006) Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J Clin Oncol 24:1665–1671
Uren AG, Kool J, Berns A et al (2005) Retroviral insertional mutagenesis: past, present and future. Oncogene 24:7656–7672
Neil JC, Cameron ER (2002) Retroviral insertion sites and cancer: fountain of all knowledge? Cancer Cell 2:253–255
Theodorou V, Kimm MA, Boer M et al (2007) MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet 39:759–769
Van Agthoven T, Van Agthoven TLA, Portengen H et al (1992) Ectopic expression of epidermal growth factor receptors induces hormone independence in ZR-75-1 human breast cancer cells. Cancer Res 52:5082–5088
Van Agthoven T, Van Agthoven TLA, Dekker A et al (1994) Induction of estrogen independence of ZR-75-1 human breast cancer cells by epigenetic alterations. Mol Endocrinol 8:1474–1483
Dorssers LCJ, Van Agthoven T, Dekker A et al (1993) Induction of antiestrogen resistance in human breast cancer cells by random insertional mutagenesis using defective retroviruses: Identification of bcar-1, a common integration site. Mol Endocrinol 7:870–878
Van Agthoven T, Van Agthoven TLA, Dekker A et al (1998) Identification of BCAR3 by a random search for genes involved in antiestrogen resistance of human breast cancer cells. EMBO J 17:2799–2808
Brinkman A, Van der Flier S, Kok EM et al (2000) BCAR1, a human homologue of the adapter protein p130Cas and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst 92:112–120
Meijer D, Van Agthoven T, Bosma PT et al (2006) Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res 4:379–386
Dorssers LCJ, Veldscholte J (1997) Identification of a novel breast-cancer-anti-estrogen-resistance (BCAR2) locus by cell-fusion-mediated gene transfer in human breast-cancer cells. Int J Cancer 72:700–705
Mikkers H, Allen J, Knipscheer P et al (2002) High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet 32:153–159
Gizard F, Robillard R, Barbier O et al (2005) TReP-132 controls cell proliferation by regulating the expression of the cyclin-dependent kinase inhibitors p21WAF1/Cip1 and p27Kip1. Mol Cell Biol 25:4335–4348
Dorssers LCJ, Van Agthoven T, Brinkman A et al (2005) Breast cancer oestrogen independence mediated by BCAR1 or BCAR3 genes is transmitted through mechanisms distinct from the oestrogen receptor signalling pathway or the epidermal growth factor receptor pathway. Breast Cancer Res 7:R82–R92. doi:10.1186/bcr1954
Dorssers LCJ, Van der Flier S, Brinkman A et al (2001) Tamoxifen resistance in breast cancer: elucidating mechanisms. Drugs 61:1721–1733
Stoica GE, Franke TF, Moroni M et al (2003) Effect of estradiol on estrogen receptor-alpha gene expression and activity can be modulated by the ErbB2/PI 3-K/Akt pathway. Oncogene 22:7998–8011
de Graffenried LA, Friedrichs WE, Russell DH et al (2004) Inhibition of mTOR activity restores tamoxifen response in breast cancer cells with aberrant Akt Activity. Clin Cancer Res 10:8059–8067
Glaros S, Atanaskova N, Zhao C et al (2006) Activation function-1 domain of estrogen receptor regulates the agonistic and antagonistic actions of tamoxifen. Mol Endocrinol 20:996–1008
Kairouz R, Parmar J, Lyons RJ et al (2005) Hormonal regulation of the Grb14 signal modulator and its role in cell cycle progression of MCF-7 human breast cancer cells. J Cell Physiol 203:85–93
O’Neill GM, Fashena SJ, Golemis EA (2000) Integrin signalling: a new cas(t) of characters enters the stage. Trends Cell Biol 10:111–119
Riggins RB, Thomas KS, Ta HQ et al (2006) Physical and functional interactions between Cas and c-Src induce tamoxifen resistance of breast cancer cells through pathways involving epidermal growth factor receptor and signal transducer and activator of transcription 5b. Cancer Res 66:7007–7015
Felekkis KN, Narsimhan RP, Near R et al (2005) AND-34 activates phosphatidylinositol 3-kinase and induces anti-estrogen resistance in a SH2 and GDP exchange factor-like domain-dependent manner. Mol Cancer Res 3:32–41
Yu Y, Hao Y, Feig LA (2006) The R-Ras GTPase mediates cross talk between estrogen and insulin signaling in breast cancer cells. Mol Cell Biol 26:6372–6380
Shou J, Massarweh S, Osborne CK et al (2004) Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst 96:926–935
Perissi V, Rosenfeld MG (2005) Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol 6:542–554
Keeton EK, Brown M (2005) Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-alpha and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol Endocrinol 19:1543–1554
Gizard F, Robillard R, Gross B et al (2006) TReP-132 is a novel progesterone receptor coactivator required for the inhibition of breast cancer cell growth and enhancement of differentiation by progesterone. Mol Cell Biol 26:7632–7644
Dressman HK, Hans C, Bild A et al (2006) Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin Cancer Res 12:819–826
Nicholson S, Halcrow P, Farndon JR et al (1989) Expression of epidermal growth factor receptors associated with lack of response to endocrine therapy in recurrent breast cancer. Lancet i:182–185
Brugge J, Hung MC, Mills GB (2007) A new mutational AKTivation in the PI3K pathway. Cancer Cell 12:104–107
Bellacosa A, De Feo D, Godwin AK et al (1995) Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer 64:280–285
Kirkegaard T, Witton CJ, McGlynn LM et al (2005) AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J Pathol 207:139–146
Van der Flier S, Brinkman A, Look MP et al (2000) Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J Natl Cancer Inst 92:120–127
Dorssers LCJ, Grebenchtchikov N, Brinkman A et al (2004) The prognostic value of BCAR1 in patients with primary breast cancer. Clin Cancer Res 10:6194–6202
Dorssers LCJ, Grebenchtchikov N, Brinkman A et al (2004) Application of a newly developed ELISA for BCAR1 Protein for prediction of clinical benefit of tamoxifen therapy in patients with advanced breast cancer. Clin Chem 50:1445–1447
Defilippi P, Di Stefano P, Cabodi S (2006) p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol 16:257–263
Cabodi S, Tinnirello A, Di Stefano P et al (2006) p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res 66:4672–4680
Kao J, Pollack JR (2006) RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes. Genes Chromosomes Cancer 45:761–769
Acknowledgements
We acknowledge Anton Berns (Amsterdam) for the splinkerette protocol, and Fred Sweep and Nicolai Grebenchtchikov (Nijmegen) for their support in generating antibodies. Excellent support of various current and former members of our research laboratory is gratefully acknowledged. For stimulating discussions and support, we are indebted to Ad Brinkman, Danielle Meijer, John Foekens, Maxime Look, Leendert Looijenga, Els Berns, John Martens, Maurice Jansen and Riccardo Fodde. Funding: This study was supported by grants of the Dutch Cancer Society (DDHK96-1245 & 99-1883), the Susan G. Komen Breast Cancer Foundation (BCTR0100675), the Association for International Cancer Research (04–148) and the Josephine Nefkens Stichting.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ton van Agthoven and Jos Veldscholte contributed equally to this research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
van Agthoven, T., Veldscholte, J., Smid, M. et al. Functional identification of genes causing estrogen independence of human breast cancer cells. Breast Cancer Res Treat 114, 23–30 (2009). https://doi.org/10.1007/s10549-008-9969-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-008-9969-5