Abstract
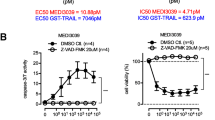
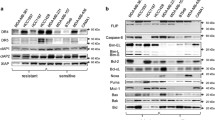
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) induces apoptosis in some but not all breast cancer cell lines. Breast cancers can be divided into those which express the estrogen (ER) and progesterone (PR) receptors, those with HER-2 amplification, and those without expression of ER, PR, or HER-2 amplification (referred to as basal or triple-negative breast cancer). We tested a panel of 20 breast cancer cell lines representing the different types of breast cancer to evaluate if the molecular phenotype of the breast cancer cells determined their response to TRAIL. The most striking finding was that eight of eleven triple-negative cell lines are sensitive to TRAIL-mediated apoptosis. The eight TRAIL-sensitive triple-negative cell lines have a mesenchymal phenotype while the three TRAIL-resistant triple-negative cell lines have an epithelial phenotype. Two of five cell lines with HER-2 amplification were sensitive to TRAIL and none of the five ER positive cell lines were sensitive. RNAi-mediated knockdown of TRAIL receptor expression demonstrated that TRAIL Receptor 2 (TRAIL-R2) mediates the effects of TRAIL, even when both TRAIL-R1 and TRAIL-R2 are expressed. Finally, inhibition of EGFR, expressed in both TRAIL-sensitive and TRAIL-resistant triple-negative breast cancer cell lines, using a small molecule tyrosine kinase inhibitor (AG1478), enhanced TRAIL-induced apoptosis in TRAIL-sensitive cell lines but did not convert resistant cells into TRAIL-sensitive cells. Together, these findings suggest that a subset of triple-negative breast cancer, those with mesenchymal features, may be the most likely to benefit from TRAIL targeted therapy. These findings could form the basis to select breast cancer patients for clinical trials of TRAIL-R2 ligands.






Similar content being viewed by others
References
Keane MM, Ettenberg SA, Nau MM, Russell EK, Lipkowitz S (1999) Chemotherapy augments TRAIL-induced apoptosis in breast cell lines. Cancer Res 59(3):734–741
Chinnaiyan A, Prasad U, Shankar S, Hamstra DA, Shanaiah M, Chenevert TL, Ross BD, Rehemtulla A (2000) Combined effect of tumor necrosis factor-related apoptosis-inducing ligand and ionizing radiation in breast cancer therapy. Proc Natl Acad Sci USA 97(4):1754–1759
Walczak H, Miller RE, Ariail K et al (1999) Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat Med 5(2):157–163
Brenton JD, Carey LA, Ahmed AA, Caldas C (2005) Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol 23(29):7350–7360
Cuello M, Ettenberg SA, Clark AS, Keane MM, Posner RH, Nau MM, Dennis PA, Lipkowitz S (2001) Down-regulation of the erbB-2 receptor by trastuzumab (herceptin) enhances tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis in breast and ovarian cancer cell lines that overexpress erbB-2. Cancer Res 61(12):4892–4900
Neve RM, Chin K, Fridlyand J et al (2006) A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10(6):515–527
Gentleman RC, Carey VJ, Bates DM et al (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5(10):R80
Smyth GK (2005) Limma: linear models for microarray data. In: Gentleman RC et al (eds) Bioinformatics and computational biology solutions using R and Bioconductor. Springer, New York, pp 397–420
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a pratical and powerful approach to multiple testing. J R Stat Soc 57:289–300
Ethier SP (1996) Human breast cancer cell lines as models of growth regulation and disease progression. J Mammary Gland Biol Neoplasia 1(1):111–121
Flanagan L, Van Weelden K, Ammerman C, Ethier SP, Welsh J (1999) SUM-159PT cells: a novel estrogen independent human breast cancer model system. Breast Cancer Res Treat 58(3):193–204
Ashkenazi A (2002) Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer 2(6):420–430
Weinberg RA (2007) The biology of cancer. Garland Science, New York
Charafe-Jauffret E, Monville F, Bertucci F et al (2007) Moesin expression is a marker of basal breast carcinomas. Int J Cancer 121(8):1779–1785
Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N, Slamon DJ (2007) Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/”triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res Treat 105(3):319–326
Korsching E, Packeisen J, Agelopoulos K et al (2002) Cytogenetic alterations and cytokeratin expression patterns in breast cancer: integrating a new model of breast differentiation into cytogenetic pathways of breast carcinogenesis. Lab Invest 82(11):1525–1533
Livasy CA, Karaca G, Nanda R, Tretiakova MS, Olopade OI, Moore DT, Perou CM (2006) Phenotypic evaluation of the basal-like subtype of invasive breast carcinoma. Mod Pathol 19(2):264–271
Gibson EM, Henson ES, Haney N, Villanueva J, Gibson SB (2002) Epidermal growth factor protects epithelial-derived cells from tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by inhibiting cytochrome c release. Cancer Res 62(2):488–496
Gibson S, Tu S, Oyer R, Anderson SM, Johnson GL (1999) Epidermal growth factor protects epithelial cells against Fas-induced apoptosis. Requirement for Akt activation. J Biol Chem 274(25):17612–17618
Park SY, Seol DW (2002) Regulation of Akt by EGF-R inhibitors, a possible mechanism of EGF-R inhibitor-enhanced TRAIL-induced apoptosis. Biochem Biophys Res Commun 295(2):515–518
Levitzki A, Gazit A (1995) Tyrosine kinase inhibition: an approach to drug development. Science 267(5205):1782–1788
Filmus J, Pollak MN, Cailleau R, Buick RN (1985) MDA-468, a human breast cancer cell line with a high number of epidermal growth factor (EGF) receptors, has an amplified EGF receptor gene and is growth inhibited by EGF. Biochem Biophys Res Commun 128(2):898–905
Lebeau J, Goubin G (1987) Amplification of the epidermal growth factor receptor gene in the BT20 breast carcinoma cell line. Int J Cancer 40(2):189–191
Charafe-Jauffret E, Ginestier C, Monville F et al (2006) Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene 25(15):2273–2284
Sorlie T, Perou CM, Tibshirani R et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98(19):10869–10874
Umemura S, Takekoshi S, Suzuki Y, Saitoh Y, Tokuda Y, Osamura RY (2005) Estrogen receptor-negative and human epidermal growth factor receptor 2-negative breast cancer tissue have the highest Ki-67 labeling index and EGFR expression: gene amplification does not contribute to EGFR expression. Oncol Rep 14(2):337–343
Willipinski-Stapelfeldt B, Riethdorf S, Assmann V, Woelfle U, Rau T, Sauter G, Heukeshoven J, Pantel K (2005) Changes in cytoskeletal protein composition indicative of an epithelial-mesenchymal transition in human micrometastatic and primary breast carcinoma cells. Clin Cancer Res 11(22):8006–8014
Hughes SC, Fehon RG (2007) Understanding ERM proteins – the awesome power of genetics finally brought to bear. Curr Opin Cell Biol 19(1):51–56
Wagner KW, Punnoose EA, Januario T et al (2007) Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med 13(9):1070–1077
Meric F, Lee WP, Sahin A, Zhang H, Kung HJ, Hung MC (2002) Expression profile of tyrosine kinases in breast cancer. Clin Cancer Res 8(2):361–367
Holland SJ, Powell MJ, Franci C et al (2005) Multiple roles for the receptor tyrosine kinase axl in tumor formation. Cancer Res 65(20):9294–9303
Evtimova V, Zeillinger R, Weidle UH (2003) Identification of genes associated with the invasive status of human mammary carcinoma cell lines by transcriptional profiling. Tumour Biol 24(4):189–198
Shipitsin M, Campbell LL, Argani P et al (2007) Molecular definition of breast tumor heterogeneity. Cancer Cell 11(3):259–273
Kelley RF, Totpal K, Lindstrom SH, Mathieu M, Billeci K, Deforge L, Pai R, Hymowitz SG, Ashkenazi A (2005) Receptor-selective mutants of apoptosis-inducing ligand 2/tumor necrosis factor-related apoptosis-inducing ligand reveal a greater contribution of death receptor (DR) 5 than DR4 to apoptosis signaling. J Biol Chem 280(3):2205–2212
MacFarlane M, Kohlhaas SL, Sutcliffe MJ, Dyer MJ, Cohen GM (2005) TRAIL receptor-selective mutants signal to apoptosis via TRAIL-R1 in primary lymphoid malignancies. Cancer Res 65(24):11265–11270
vander Sloot AM, Tur V, Szegezdi E, Mullally MM, Cool RH, Samali A, Serrano L, Quax WJ (2006) Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci USA 103(23):8634–8639
Acknowledgements
We thank Steven Shaw for helpful discussion about moesin and for providing the anti-moesin antibody. We thank Jeffrey Rubin for his careful review of this manuscript and thoughtful comments. Funding This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Additional information
S. R. Davis and J. G. Pumphrey contributed equally to this work.
Electronic supplementary material
Table S1
List of genes with a false discovery rate of 2% for the regression analysis of TRAIL sensitivity and gene expression ranked from most to least significant. Column headings are, from left to right, official gene symbol, gene description, Entrez Gene ID, Affymetrix probeset ID, t-statistic, and false discovery rate (adj.P.val). A positive t-statistic is associated with genes that are higher in TRAIL sensitive cell lines. The raw expression data described in Neve et al. [6] was downloaded from http://caArray.nci.nih.gov. (PDF 23 kb)
Rights and permissions
About this article
Cite this article
Rahman, M., Davis, S.R., Pumphrey, J.G. et al. TRAIL induces apoptosis in triple-negative breast cancer cells with a mesenchymal phenotype. Breast Cancer Res Treat 113, 217–230 (2009). https://doi.org/10.1007/s10549-008-9924-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-008-9924-5