Abstract
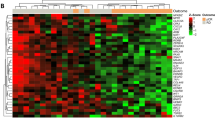
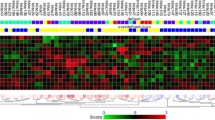
Cyclophosphamide, methotrexate and 5-fluorouracile (CMF)-based chemotherapy for adjuvant treatment of breast cancer reduces the risk of relapse. In this exploratory study, we tested the feasibility of identifying molecular markers of recurrence in CMF-treated patients. Using Affymetrix U133A GeneChips, RNA samples from 19 patients with primary breast cancer who had been uniformly treated with adjuvant CMF chemotherapy were analyzed. Two supervised class prediction approaches were used to identify gene markers that can best discriminate between patients who would experience relapse and patients who would remain disease-free. An additional independent validation set of 51 patients and 21 genes were analyzed by quantitative RT-PCR. Applying different algorithms to evaluate our microarray data, we identified two gene expression signatures of 21 and 12 genes containing eight overlapping genes, that predict recurrence in 19 cases with high accuracy (94%). Quantitative RT-PCR demonstrated that six genes from the combined signatures (CXCL9, ITSN2, GNAI2, H2AFX, INDO, and MGC10986) were significantly differentially expressed in the recurrence versus the non-recurrence group of the 19 cases and the independent breast cancer patient cohort (n = 51) treated with CMF. High expression levels of CXCL9, ITSN2, and GNAI2 were associated with prolonged disease-free survival (DFS) (P = 0.029, 0.018 and 0.032, respectively). When patients were stratified by combined CXCL9/ITSN2 or CXCL9/FLJ22028 tumor levels, they exhibited significantly different disease-free survival curves (P = 0.0073 and P = 0.005, respectively). Finally, the CXCL9/ITSN2 and CXCL9/FLJ22028 ratio was an independent prognostic factor (P = 0.034 and P = 0.003, respectively) for DFS by multivariate Cox analysis in the 70-patient cohort. Our data highlight the feasibility of a prognostic assay that is applicable to therapeutic decision-making for breast cancer. Whether the biomarker profile is chemotherapy-specific or whether it is a more general indicator of bad prognosis of breast cancer patients remains to be explored.



Similar content being viewed by others
References
Eifel P, Axelson JA, Costa J, Crowley J, Curran WJ Jr, Deshler A et al (2001) National Institutes of Health Consensus Development Conference Statement: adjuvant therapy for breast cancer, November 1–3, 2000. J Natl Cancer Inst 93:979–989. doi:10.1093/jnci/93.13.979
Early Breast Cancer Trialists’ Collaborative Group (1992) Systemic treatment of early breast cancer by hormonal, cytotoxic, or immune therapy. 133 randomised trials involving 31,000 recurrences and 24,000 deaths among 75,000 women. Lancet 339:1–15
Early Breast Cancer Trialists’ Collaborative Group (1998) Polychemotherapy for early breast cancer: an overview of the randomised trials. Lancet 352:930–942. doi:10.1016/S0140-6736(98)03301-7
Bonadonna G, Valagussa P, Moliterni A, Zambetti M, Brambilla C (1995) Adjuvant cyclophosphamide, methotrexate, and fluorouracil in node-positive breast cancer: the results of 20 years of follow-up. N Engl J Med 332:901–906. doi:10.1056/NEJM199504063321401
Levine MN, Bramwell VH, Pritchard KI, Norris BD, Shepherd LE, Abu-Zahra H et al (1998) Randomized trial of intensive cyclophosphamide, epirubicin, and fluorouracil chemotherapy compared with cyclophosphamide, methotrexate, and fluorouracil in premenopausal women with node-positive breast cancer. National Cancer Institute of Canada Clinical Trials Group. J Clin Oncol 16:2651–2658
Goldhirsch A, Glick JH, Gelber RD, Coates AS, Thurlimann B, Senn HJ (2005) Meeting highlights: international expert consensus on the primary therapy of early breast cancer 2005. Ann Oncol 16:1569–1583. doi:10.1093/annonc/mdi326
Pritchard KI, Shepherd LE, O’Malley FP, Andrulis IL, Tu D, Bramwell VH et al (2006) HER2 and responsiveness of breast cancer to adjuvant chemotherapy. N Engl J Med 354:2103–2111. doi:10.1056/NEJMoa054504
Askmalm MS, Carstensen J, Nordenskjold B, Olsson B, Rutqvist LE, Skoog L et al (2004) Mutation and accumulation of p53 related to results of adjuvant therapy of postmenopausal breast cancer patients. Acta Oncol 43:235–244. doi:10.1080/02841860410029474
Andersson J, Larsson L, Klaar S, Holmberg L, Nilsson J, Inganas M et al (2005) Worse survival for TP53 (p53)-mutated breast cancer patients receiving adjuvant CMF. Ann Oncol 16:743–748. doi:10.1093/annonc/mdi150
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M et al (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826. doi:10.1056/NEJMoa041588
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19:403–410. doi:10.1111/j.1365-2559.1991.tb00229.x
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286:531–537. doi:10.1126/science.286.5439.531
Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E et al (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA 96:2907–2912. doi:10.1073/pnas.96.6.2907
van‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415:530–536. doi:10.1038/415530a
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW et al (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347:1999–2009. doi:10.1056/NEJMoa021967
Hwang D, Schmitt WA, Stephanopoulos G (2002) Determination of minimum sample size and discriminatory expression patterns in microarray data. Bioinformatics 18:1184–119317
Mukherjee S, Tamayo P, Rogers S, Rifkin R, Engle A, Campbell C et al (2003) Estimating dataset size requirements for classifying DNA microarray data. J Comput Biol 10:119–142
Ayers M, Symmans WF, Stec J, Damokosh AI, Clark E, Hess K et al (2004) Gene expression profiles predict complete pathologic response to neoadjuvant paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide chemotherapy in breast cancer. J Clin Oncol 22:2284–2293. doi:10.1200/JCO.2004.05.166
Datta S, Datta S (2003) Comparisons and validation of statistical clustering techniques for microarray gene expression data. Bioinformatics 19:459–466. doi:10.1093/bioinformatics/btg025
Hilsenbeck SG, Friedrichs WE, Schiff R, O’Connell P, Hansen RK, Osborne CK et al (1999) Statistical analysis of array expression data as applied to the problem of tamoxifen resistance. J Natl Cancer Inst 91:453–459. doi:10.1093/jnci/91.5.453
Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF et al (2003) Gene expression predictors of breast cancer outcomes. Lancet 361:1590–1596. doi:10.1016/S0140-6736(03)13308-9
Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A et al (2003) Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA 100:10393–10398. doi:10.1073/pnas.1732912100
Sgadari C, Farber JM, Angiolillo AL, Liao F, Teruya-Feldstein J, Burd PR et al (1997) Mig, the monokine induced by interferon-gamma, promotes tumor necrosis in vivo. Blood 89:2635–2643
Dorsey R, Kundu N, Yang Q, Tannenbaum CS, Sun H, Hamilton TA et al (2002) Immunotherapy with interleukin-10 depends on the CXC chemokines inducible protein-10 and monokine induced by IFN-gamma. Cancer Res 62:2606–2610
Kondo T, Ito F, Nakazawa H, Horita S, Osaka Y, Toma H (2004) High expression of chemokine gene as a favorable prognostic factor in renal cell carcinoma. J Urol 171:2171–2175. doi:10.1097/01.ju.0000127726.25609.87
Mochizuki N, Ohba Y, Kiyokawa E, Kurata T, Murakami T, Ozaki T et al (1999) Activation of the ERK/MAPK pathway by an isoform of rap1GAP associated with G alpha(i). Nature 400:891–894. doi:10.1038/23738
Pucharcos C, Estivill X, de la Luna S (2000) Intersectin 2, a new multimodular protein involved in clathrin-mediated endocytosis. FEBS Lett 478:43–51. doi:10.1016/S0014-5793(00)01793-2
Schafer DA (2002) Coupling actin dynamics and membrane dynamics during endocytosis. Curr Opin Cell Biol 14:76–81. doi:10.1016/S0955-0674(01)00297-6
Niméus-Malmström E, Ritz C, Edén P, Johnsson A, Ohlsson M, Strand C et al (2006) Gene expression profiles and conventional clinical markers to predict distant recurrences for premenopausal breast cancer patients after adjuvant chemotherapy. Eur J Cancer 42:2729–2737. doi:10.1016/j.ejca.2006.06.031
Paik S, Tang G, Shak S, Kim C, Baker J, Kim W et al (2006) Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 24:3726–3734. doi:10.1200/JCO.2005.04.7985
Martin M, Villar A, Sole-Calvo A, Gonzalez R, Massuti B, Lizon J et al (2003) Doxorubicin in combination with fluorouracil and cyclophosphamide (i.v. FAC regimen, day 1, 21) versus methotrexate in combination with fluorouracil and cyclophosphamide (i.v. CMF regimen, day 1, 21) as adjuvant chemotherapy for operable breast cancer: a study by the GEICAM group. Ann Oncol 14:833–842. doi:10.1093/annonc/mdg260
Bonadonna G, Zambetti M, Moliterni A, Gianni L, Valagussa P (2004) Clinical relevance of different sequencing of doxorubicin and cyclophosphamide, methotrexate, and fluorouracil in operable breast cancer. J Clin Oncol 22:1614–1620. doi:10.1200/JCO.2004.07.190
Chang JC, Wooten EC, Tsimelzon A, Hilsenbeck SG, Gutierrez MC, Elledge R et al (2003) Gene expression profiling for the prediction of therapeutic response to docetaxel in patients with breast cancer. Lancet 362:362–369. doi:10.1016/S0140-6736(03)14023-8
Thuerigen O, Schneeweiss A, Toedt G, Warnat P, Hahn M, Kramer H et al (2006) Gene expression signature predicting pathologic complete response with gemcitabine, epirubicin, and docetaxel in primary breast cancer. J Clin Oncol 24:1839–1845. doi:10.1200/JCO.2005.04.7019
Rody A, Karn T, Gätje R, Kourtis K, Minckwitz G, Loibl S et al (2006) Gene expression profiles of breast cancer obtained from core cut biopsies before neoadjuvant docetaxel, adriamycin, and cyclophoshamide chemotherapy correlate with routine prognostic markers and could be used to identify predictive signatures. Zentralbl Gynakol 128:76–81. doi:10.1055/s-2006-921508
Rody A, Karn T, Solbach C, Gaetje R, Munnes M, Kissler S et al (2007) The erbB2+ cluster of the intrinsic gene set predicts tumor response of breast cancer patients receiving neoadjuvant chemotherapy with docetaxel, doxorubicin and cyclophosphamide within the GEPARTRIO trial. Breast 16:235–240. doi:10.1016/j.breast.2007.02.006
Acknowledgments
Supported by a grant to M. Kiechle and H. Hoefler from the BMBF (Federal Ministry of Education and Research), Germany, German National Genome Project (KR-S15T03).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Specht, K., Harbeck, N., Smida, J. et al. Expression profiling identifies genes that predict recurrence of breast cancer after adjuvant CMF-based chemotherapy. Breast Cancer Res Treat 118, 45–56 (2009). https://doi.org/10.1007/s10549-008-0207-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-008-0207-y