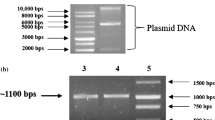
Abstract
Bacillus flexus strain SSAI1 isolated from agro-industry waste, Tuem, Goa, India displayed high arsenite resistance as minimal inhibitory concentration was 25 mM in mineral salts medium. This bacterial strain exposed to 10 mM arsenite demonstrated rapid arsenite oxidation and internalization of 7 mM arsenate within 24 h. The Fourier transformed infrared (FTIR) spectroscopy of cells exposed to arsenite revealed important functional groups on the cell surface interacting with arsenite. Furthermore, scanning electron microscopy combined with electron dispersive X-ray spectroscopy (SEM-EDAX) of cells exposed to arsenite revealed clumping of cells with no surface adsorption of arsenite. Transmission electron microscopy coupled with electron dispersive X-ray spectroscopic (TEM-EDAX) analysis of arsenite exposed cells clearly demonstrated ultra-structural changes and intracellular accumulation of arsenic. Whole-genome sequence analysis of this bacterial strain interestingly revealed the presence of large number of metal(loid) resistance genes, including aioAB genes encoding arsenite oxidase responsible for the oxidation of highly toxic arsenite to less toxic arsenate. Enzyme assay further confirmed that arsenite oxidase is a periplasmic enzyme. The genome of strain SSAI1 also carried glpF, aioS and aioE genes conferring resistance to arsenite. Therefore, multi-metal(loid) resistant arsenite oxidizing Bacillus flexus strain SSAI1 has potential to bioremediate arsenite contaminated environmental sites and is the first report of its kind.






Similar content being viewed by others
References
Aguilar NC, Faria MC, Pedron T, Batista BL, Mesquita JP, Bomfeti CA, Rodrigues JL (2020) Isolation and characterization of bacteria from a brazilian gold mining area with a capacity of arsenic bioaccumulation. Chemosphere 240:124871
Anderson GL, Williams J, Hille R (1992) The purification and characterization of arsenite oxidase from Alcaligenes faecalis, a molybdenum-containing hydroxylase. J Biol Chem 267:23674–23682
Arsène-Ploetze F, Koechler S, Marchal M, Coppée JY, Chandler M, Bonnefoy V, Brochier-Armanet C, Barakat M, Barbe V, Battaglia-Brunet F, Bruneel O (2010) Structure, function, and evolution of the Thiomonas spp. genome. PLoS Genet 6(2):e1000859
Bagade A, Nandre V, Paul D, Patil Y, Sharma N, Giri A, Kodam K (2020) Characterisation of hyper tolerant Bacillus firmus L- 148 for arsenic oxidation. Environ Pollut 261: 114124.
Bahar MM, Megharaj M, Naidu R (2012) Arsenic bioremediation potential of a new arsenite oxidizing bacterium Stenotrophomonas sp. MM-7 isolated from soil. Biodegradation 23(6):803–812
Bahar MM, Megharaj M, Naidu R (2013) Kinetics of arsenite oxidation by Variovorax sp. MM-1 isolated from a soil and identification of arsenite oxidase gene. J Hazard Mater 262:997–1003
Banerjee S, Datta S, Chattyopadhyay D, Sarkar P (2011) Arsenic accumulating and transforming bacteria isolated from contaminated soil for potential use in bioremediation. J Environ Sci Health Part A 46(14):1736–1747
Bermanec V, Paradžik T, Kazazić SP, Venter C, Hrenović J, Vujaklija D, Duran R, Boev I, Boev B (2020) Novel arsenic hyper-resistant bacteria from an extreme environment, Crven Dol mine, Allchar, North Macedonia. J Hazard Mater 402:123437
Branco R, Francisco R, Chung AP, Morais PV (2009) Identification of an aox system that requires cytochrome c in the highly arsenic-resistant bacterium Ochrobactrum tritici SCII24. Appl Environ Microbiol 75(15):5141–5147
Bueno BYM, Torem ML, Molina FALMS, De Mesquita LMS (2008) Biosorption of lead(II), chromium(III) and copper(II) by R. opacus: equilibrium and kinetic studies. Miner Eng 21(1):65–75
Cai L, Liu G, Rensing C, Wang G (2009a) Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol 9(1):4
Cai L, Rensing C, Li X, Wang G (2009b) Novel gene clusters involved in arsenite oxidation and resistance in two arsenite oxidizers: Achromobacter sp. SY8 and Pseudomonas sp. TS44. Appl Microbiol Biotechnol 83(4):715–725
Chang JS (2015) Biotransformation of arsenite and bacterial aox activity in drinking water produced from surface water of floating houses: arsenic contamination in Cambodia. Environ Pollut 206:315–323
Chang JS, Yoon IH, Lee JH, Kim KR, An J, Kim KW (2010) Arsenic detoxification potential of aox genes in arsenite-oxidizing bacteria isolated from natural and constructed wetlands in the Republic of Korea. Environ Geochem Health 32(2):95–105
Dey U, Chatterjee S, Mondal NK (2016) Isolation and characterization of arsenic-resistant bacteria and possible application in bioremediation. Biotechnol Rep 10:1–7
Goswami R, Mukherjee S, Rana VS, Saha DR, Raman R, Padhy PK, Mazumder S (2015) Isolation and characterization of arsenic-resistant bacteria from contaminated water-bodies in West Bengal, India. Geomicrobiol J 32(1):17–26
Gourbal B, Sonuc N, Bhattacharjee H, Legare D, Sundar S, Ouellette M, Rosen BP, Mukhopadhyay R (2004) Drug uptake and modulation of drug resistance in Leishmania by an aquaglyceroporin. J Biol Chem 279:31010–31017
Guo H, Liu Z, Ding S, Hao C, Xiu W, Hou W (2015) Arsenate reduction and mobilization in the presence of indigenous aerobic bacteria obtained from high arsenic aquifers of the Hetao basin, Inner Mongolia. Environ Pollut 203:50–59
Jain R, Jha S, Adhikary H, Kumar P, Parekh V, Jha A, Mahatma MK, Kumar GN (2014) Isolation and molecular characterization of arsenite-tolerant Alishewanella sp. GIDC-5 originated from industrial effluents. Geomicrobiol J 31(1):82–90
Jebeli MA, Maleki A, Amoozegar MA, Kalantar E, Izanloo H, Gharibi F (2017) Bacillus flexus strain As-12, a new arsenic transformer bacterium isolated from contaminated water resources. Chemosphere 169:636–641
Jebelli MA, Maleki A, Amoozegar MA, Kalantar E, Gharibi F, Darvish N, Tashayoe H (2018) Isolation and identification of the native population bacteria for bioremediation of high levels of arsenic from water resources. J Environ Manage 212:39–45
Jia MR, Tang N, Cao Y, Chen Y, Han YH, Ma LQ (2019) Efficient arsenate reduction by As-resistant bacterium Bacillus sp. strain PVR-YHB1-1: characterization and genome analysis. Chemosphere 218:1061–1070
Kang YS, Bothner B, Rensing C, McDermott TR (2012) Involvement of RpoN in regulating bacterial arsenite oxidation. Appl Environ Microbiol 78(16):5638–5645
Kashyap DR, Botero LM, Franck WL, Hassett DJ, McDermott TR (2006) Complex regulation of arsenite oxidation in Agrobacterium tumefaciens. J Bacteriol 188(3):1081–1088
Khairul I, Wang QQ, Jiang YH, Wang C, Naranmandura H (2017) Metabolism, toxicity and anticancer activities of arsenic compounds. Oncotarget 8(14):23905–23926
Koechler S, Cleiss-Arnold J, Proux C, Sismeiro O, Dillies MA, Goulhen-Chollet F, Hommais F, Lièvremont D, Arsène-Ploetze F, Coppée JY, Bertin PN (2010) Multiple controls affect arsenite oxidase gene expression in Herminiimonas arsenicoxydans. BMC Microbiol 10(1):53
Koechler S, Arsène-Ploetze F, Brochier-Armanet C, Goulhen-Chollet F, Heinrich-Salmeron A, Jost B, Lièvremont D, Philipps M, Plewniak F, Bertin PN, Lett MC (2015) Constitutive arsenite oxidase expression detected in arsenic-hypertolerant Pseudomonas xanthomarina S11. Res Microbiol 166(3):205–214
Kruger MC, Bertin PN, Heipieper HJ, Arsène-Ploetze F (2013) Bacterial metabolism of environmental arsenic—mechanisms and biotechnological applications. Appl Microbiol Biotechnol 97(9):3827–3841
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lett M, Paknikar K, Lievremont D (2001) A simple and rapid method for arsenite and arsenate speciation. Process Metall 11:541–546
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1):265–275
Mahtani S, Mavinkurve S (1979) Microbial purification of longifolene: a sesquiterpene. J Ferment Technol 57:529–533
Majumder A, Bhattacharyya K, Bhattacharyya S, Kole SC (2013) Arsenic-tolerant, arsenite-oxidising bacterial strains in the contaminated soils of West Bengal, India. Sci Total Environ 463:1006–1014
Mallick I, Hossain ST, Sinha S, Mukherjee SK (2014) Brevibacillussp. KUMAs2, a bacterial isolate for possible bioremediation of arsenic in rhizosphere. Ecotoxicol Environ Saf 107:236–244
Mallick I, Bhattacharyya C, Mukherji S, Dey D, Sarkar SC, Mukhopadhyay UK, Ghosh A (2018) Effective rhizoinoculation and biofilm formation by arsenic immobilizing halophilic plant growth promoting bacteria (PGPB) isolated from mangrove rhizosphere: a step towards arsenic rhizoremediation. Sci Total Environ 610:1239–1250
Mandal BK, Suzuki KT (2002) Arsenic round the world: a review. Talanta 58(1):201–235
Meng YL, Liu Z, Rosen BP (2004) As(III) and Sb(III) uptake by GlpF and efflux by ArsB in Escherichia coli. J Biol Chem 279(18):18334–18341
Mujawar SY, Shamim K, Vaigankar DC, Dubey SK (2019) Arsenite biotransformation and bioaccumulation by Klebsiella pneumoniae strain SSSW7 possessing arsenite oxidase (aioA) gene. Biometals 32(1):65–76
Muller D, Médigue C, Koechler S, Barbe V, Barakat M, Talla E, Bonnefoy V, Krin E, Arsene-Ploetze F, Carapito C, Chandler M (2007) A tale of two oxidation states: bacterial colonization of arsenic-rich environments. PLoS Genet 3(4):e53
Naumann D, Helm D, Labischinski H, Giesbrecht P (1991) The characterization of microorganisms by Fourier-transform infrared spectroscopy. In: Nelson WH (ed) Modern techniques for rapid microbiological analysis. VCH, New York, pp 43–96
Oust A, Møretrø T, Kirschner C, Narvhus JA, Kohler A (2004) FT-IR spectroscopy for identification of closely related Lactobacilli. J Microbiol Methods 59(2):49–162
Páez-Espino D, Tamames J, de Lorenzo V, Cánovas D (2009) Microbial responses to environmental arsenic. Biometals 22(1):117–130
Palma-Lara I, Martínez-Castillo M, Quintana-Pérez JC, Arellano-Mendoza MG, Tamay-Cach F, Valenzuela-Limón OL, García-Montalvo EA, Hernández-Zavala A (2020) Arsenic exposure: a public health problem leading to several cancers. Regul Toxicol Pharmacol 110:104539
Pandi M, Shashirekha V, Swamy M (2009) Bioabsorption of chromium from retan chrome liquor by cyanobacteria. Microbiol Res 164(4):420–428
Rahman MM, Ng JC, Naidu R (2009) Chronic exposure of arsenic via drinking water and its adverse health impacts on humans. Environ Geochem Health 31:189–200
Rathod J, Dhanani AS, Jean JS, Jiang WT (2019) The whole genome insight on condition-specific redox activity and arsenopyrite interaction promoting As-mobilization by strain Lysinibacillus sp. B2A1. J Hazard Mater 364:671–681
Rehman A, Butt SA, Hasnain S (2010) Isolation and characterization of arsenite oxidizing Pseudomonas lubricans and its potential use in bioremediation of wastewater. Afr J Biotechnol 9(10):1493–1498
Santini JM, van den Hoven RN (2004) Molybdenum-containing arsenite oxidase of the chemolithoautotrophic arsenite oxidizer NT-26. J Bacteriol 186:1614–1619
Sardiwal S, Santini JM, Osborne TH, Djordjevic S (2010) Characterization of a two-component signal transduction system that controls arsenite oxidation in the chemolithoautotroph NT-26. FEMS Microbiol Lett 313(1):20–28
Satyapal GK, Rani S, Kumar M, Kumar N (2016) Potential role of arsenic resistant bacteria in bioremediation: current status and future prospects. J Microb Biochem Technol 8(3):256–258
Saunders JK, Rocap G (2016) Genomic potential for arsenic efflux and methylation varies among global Prochlorococcus populations. ISME J 10(1):197–209
Singh N, Gupta S, Marwa N, Pandey V, Verma PC, Rathaur S, Singh N (2016) Arsenic mediated modifications in Bacillus aryabhattai and their biotechnological applications for arsenic bioremediation. Chemosphere 164:524–534
Sodhi KK, Kumar M, Agrawal PK, Singh DK (2019) Perspectives on arsenic toxicity, carcinogenicity and its systemic remediation strategies. Environ Technol Innov 16:100462
Studholme DJ, Jackson RA, Leak DJ (1999) Phylogenetic analysis of transformable strains of thermophilic Bacillus species. FEMS Microbiol Lett 172:85–90
Thomas DJ, Rosen BP (2013) Arsenic methyltransferases. In: Kretsinger RH, Uversky VN, Permyakov EA (eds) Encyclopedia of metalloproteins. Springer, New York, pp 140–145
van den Hoven RN, Santini JM (2004) Arsenite oxidation by the heterotroph Hydrogenophaga sp. strain NT-14: the arsenite oxidase and its physiological electron acceptor. Biochim Biophys Acta Bioenergetics 1656(2–3):148–155
Wang Q, Han Y, Shi K, Fan X, Wang L, Li M, Wang G (2017) An oxidoreductase AioE is responsible for bacterial arsenite oxidation and resistance. Sci Rep 7(1):1–10
Wu Y, Feng S, Li B, Mi X (2010) The characteristics of Escherichia coli adsorption of arsenic(III) from aqueous solution. World J Microbiol Biotechnol 26(2):249–256
Zhao FJ, McGrath SP, Meharg AA (2010) Arsenic as a food chain contaminant: mechanisms of plant uptake and metabolism and mitigation strategies. Ann Rev Plant Biol 61:535–559
Acknowledgements
Ms. Sajiya Yusuf Mujawar is grateful to University Grants Commission, New Delhi for financial support as MANF-SRF (Award Letter No. F1-17.1/2015-16/MANF-2015-17-Goa-50641). The authors are thankful to Mr. Areef Sardar from CSIR-National Institute of Oceanography, Goa for EDAX analysis and AIRF, Jawaharlal Nehru University, New Delhi for TEM-EDAX analysis. The authors are also obliged to Dr. B. R. Srinivasan and Mr. Rahul Kerkar from Department of Chemistry, Goa University for FTIR analysis. The authors are also grateful to Dr. Sandeep Garg, Head, Department of Microbiology, Goa University for providing laboratory facilities.
Author information
Authors and Affiliations
Contributions
SYM: Conceptualization, experiments, data generation, analysis and writing of the original draft manuscript. DCV: Assisted in experimental work and draft writing. SKD: Experimental designs, verification of data, mentoring of experiments, correction and final editing of the research manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mujawar, S.Y., Vaigankar, D.C. & Dubey, S.K. Biological characterization of Bacillus flexus strain SSAI1 transforming highly toxic arsenite to less toxic arsenate mediated by periplasmic arsenite oxidase enzyme encoded by aioAB genes. Biometals 34, 895–907 (2021). https://doi.org/10.1007/s10534-021-00316-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10534-021-00316-x