Abstract
Marine organisms that enter the Mediterranean from the Red Sea via the Suez Canal are known as Lessepsian bioinvaders. In general, genetic studies of Lessepsian fishes have shown little structure between Red Sea and Mediterranean populations. Yet notable exceptions suggest the importance of life-history factors that may influence patterns of spatial genetic variation. In this study, by sampling two invasive fishes with different life histories (the rabbitfish Siganus rivulatus and the filefish Stephanolepis diaspros), we looked at evidence of population structure and selection at the boundary between the Red Sea and the Mediterranean (the Suez Canal), using thousands of molecular markers. Results illustrate two divergent patterns of genetic patterns, with little genetic structure in S. rivulatus and strong population structure in S. diaspros, even at such small spatial scale. We discuss differences in ecological characteristics between the two species to account for such differences. In addition, we report that in the face of both high (S. rivulatus) and low (S. diaspros) gene flow, loci under selection were uncovered, and some protein coding genes were identified as being involved with osmoregulation, which seems to be an important feature of individuals crossing the salinity-variable Suez Canal. The presence of genes under selection in populations near the Suez Canal supports the idea that selection may be active and essential for successful invasions right out of the gate.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Over the last decades, molecular techniques have provided important innovation in the field of invasion biology (Rius et al. 2015), especially in the last few years with the use of genomic approaches (Bernardi et al. 2016; Barbosa 2021; Stuart et al. 2021). Particularly, population genetics and phylogenetics are increasingly used to investigate some key aspects of biological invasions, such as the dynamic patterns of genetic diversity and evolutionary processes in real time (Dlugosch and Parker 2008; Blackwell et al. 2020; Popovic and Bernatchez 2021).
Introductions may result from the transport of few individuals to a new location where they thrive, in part due to being released from predation, competition and parasitic load pressures (Baker and Stebbins 1965; Sax et al. 2007). For example, the recent explosion of lionfishes in the Caribbean is a spectacular illustration of the devastating effects of biological invasions (Betancur et al. 2011). While apparently straightforward, the genetics of bioinvasions are counterintuitive (Dlugosch and Parker 2008), as shown by empirical studies that oppose general predictions (Chiesa et al. 2019). Theoretically, founding effects should be revealed by a decrease in genetic diversity resulting from bottlenecked populations. Nevertheless, despite their lowered evolutionary potential, and due to a combination of ecological and biological characteristics, these species can still become successful in their new environment (e.g. Azzurro et al. 2006; Xue et al. 2018). This is known, in invasion biology, as the ‘genetic paradox’ (Roman and Darling 2007).
In other cases, and especially in aquatic systems (e.g. Holland 2001; Xue et al. 2018), invasive populations may maintain a high genetic diversity with no trace of bottleneck. This situation seems to be very common in the Mediterranean Sea (e.g. Bernardi et al. 2010), one of the most invaded region of the world, with recent estimates of the number of introduced species close to 700 established metazoans (Zenetos and Galanidi 2020).
In 1869, the opening of the Suez Canal, started a process of invasion from the Red Sea into the Mediterranean (termed Lessepsian migration, after the Canal engineer Ferdinand de Lesseps), resulting in a variety of ecological impacts on the receiving ecosystem (Por 1978; Golani 2010; Katsanevakis et al. 2016). Whilst species moving in the opposite direction, the so-called ‘anti-Lessepsian migrants’, are very rare (Tortonese 1984; Bos et al. 2020). Lessepsian fishes now comprise more than 100 species (Zenetos et al. 2012; Azzurro et al. 2015), and they are increasingly the focus of genetic studies (Chiesa et al. 2019). In general, the specific dynamics of biological invasions are poorly known, yet, in this case, the situation presents some definite advantages for scientists. The date of the opening of the invasion route is known, and the geographical source of the invaders is also known, at least in general terms, as the Red Sea region. Thus, theoretical predictions seem fairly simple. Some individuals from the Red Sea would enter the Mediterranean via the Suez Canal and would later expand into the wide-open ecological niches. This situation would predict a genetic bottleneck due to an invading subsample of the original populations, followed by a fast range expansion, a pattern that is consistent with other documented invasions (Sax et al. 2005). However, in contrast to this prediction, Red Sea and invasive Mediterranean populations studied so far displayed a high genetic similarity and no evidence of genetic bottleneck (Golani and Ritte 1999; Bucciarelli et al. 2002; Hassan et al. 2003; Hassan and Bonhomme 2005; Azzurro et al. 2006; Bariche and Bernardi 2009), with notable exceptions such as the bluespotted cornetfish, Fistularia commersonii (Golani et al. 2007; Tenggardjaja et al. 2014; Jackson et al. 2015; Bernardi et al. 2016). In general, data suggest that colonization occurred by a large number of individuals or by multiple colonization events (Chiesa et al. 2019).
Possible explanations for the genetic patterns of Lessepsian fishes are that introductions are only successful when a large number of individuals enter the Mediterranean at once, (or that multiple introduction events occur in rapid succession). A further hypothesis is that individuals undergo preadaptation (genetic changes prior to introduction), and when selected individuals enter the new environment, these changes become adaptive and the invasion successful (Blackburn et al. 2017; Stuart et al. 2020; Barbosa 2021). Key components missing to our understanding of the Lessepsian bioinvasions are the temporal and spatial dimensions of the earliest phase of invasion. So far, only two studies assessed the temporal component: blue-chin parrotfish, Scarus ghobban, individuals were collected in Lebanon very soon after first being detected, and so were dusky spinefoot rabbitfish Siganus luridus, as soon as they reached the island of Linosa, Italy. Yet, in both cases, no evidence of bottleneck was observed (Azzurro et al. 2006; Bariche and Bernardi 2009).
In this study, we focused our attention on the spatial component of the question, by looking at the region closest to the Suez Canal, in order to determine if any evidence of population structure or selection was present at the boundary between the Red Sea and the Mediterranean. We used two species of reef fishes: a rabbitfish, the marbled spinefoot Siganus rivulatus, and a filefish, the reticulated leatherjacket, Stephanolepis diaspros, to investigate the potential link between population structure and natural selection on invasion success using thousands of Single Nucleotide Polymorphisms (SNPs) obtained by RAD sequencing.
The goal of this study was, therefore, to sample these two reef fishes with different life histories in the vicinity of the Suez Canal, to assess the spatial component of Lessepsian bioinvasions at the entry gate of the Mediterranean, using thousands of molecular markers to assess the relative role of drift and selection in their invasion success.
Materials and methods
Siganus rivulatus and Stephanolepis diaspros
The two focal species have similar invasion histories, being among the first Lessepsian immigrants to having been observed in the Mediterranean in 1927 (Steinitz 1927), and to establish permanent populations in the Levant. In the last decades, these two species have significantly expanded their distribution westwards. The marbled spinefoot had reached the Adriatic coast by the year 2000 (Pallaoro and Dulc 2004), and the Strait of Sicily by 2017 (Stamouli et al. 2017). The reticulated leatherjacket had reached Tunisian, Maltese, and Italian waters by 2015, (Deidun et al. 2015; Tiralongo et al. 2019; Katsanevakis et al. 2020). The two species have, however, different life histories. Both species produce benthic adhesive eggs (Lam 1974; Popper et al. 1979), however, after hatching, pelagic larval duration differs. On the Lebanese coast, spawning occurs in June in S. rivulatus (Bariche et al. 2004), which may indicate a pelagic larval life duration of approximately 30 days, and thus fits the usual time of larval life recorded in this family (Bryan and Madraisau 1977; Popper et al. 1979). In contrast, pelagic larval duration is restricted to only 17–20 days in Stephanolepis (Rogers et al. 2001). The rabbitfish periodically spawn very large egg masses (average female carries over 200,000 oocytes), and display a ‘group-synchronous’ strategy of ovarian development (Bariche et al. 2009), while Stephanolepis have parental egg care and spawn comparatively fewer eggs (average female carries approximately 50,000 oocytes) (Mancera-Rodríguez and Castro-Hernández 2015).
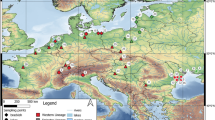
Sampling Samples of Siganus rivulatus and Stephanolepis diaspros were collected by local fishers, and with beach seines and hand nets in June and July 2010. Samples were collected in the vicinity of the Suez Canal, within 50 km of the Canal in the Red Sea, and within 300 km of the Canal in the Mediterranean. A total of 38 and 40 samples of S. rivulatus and S. diaspros, respectively, were collected in Egypt and Israel, with an additional nine samples of S. diaspros being collected in Tunisia to evaluate longer range spatial effects (Fig. 1, Table 1). Fin clips were immediately placed in 95% ethanol and kept at ambient temperature until reaching the laboratory where they were stored at − 20 °C.
Map of the study region showing sampling locations used in this study: HAI: Haifa, Israel; PSD: Port Said, Egypt; ISM: Ismailia, Egypt, SUE: Suez, Egypt; ATT: Attaka, Egypt; SOK: Sokhna, Egypt; ALE: Alexandria, Egypt; DJE: Djerba, Tunisia. Blue dots indicate sampling locations for Siganus rivulatus sampling locations, red dots indicate sampling locations for Stephanolepis diaspros, purple dots indicate sampling locations for both species
DNA extraction—Library preparation DNA was extracted from fin clips using DNeasy Blood & Tissue kits (Qiagen) according to the manufacturer's protocol. We constructed RAD libraries using a variation of the original protocol with the restriction enzyme SbfI (Miller et al. 2007, 2012; Baird et al. 2008; Longo and Bernardi 2015). Initial genomic DNA amount for each individual was 400 ng. Libraries were physically sheared on a Covaris S2 sonicator with an intensity of 5, duty cycle of 10%, cycles/burst of 200 and a cycle time of 30 s. We carried out the final PCR amplification step in 50 μl reaction volumes with 10 amplification cycles. Ampure XP beads (Agencourt) were used for each purification step and size selection. The library was sequenced in a single lane on an Illumina HiSeq 4000 at the Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley (USA). Then, we applied Perl scripts to trim the raw reads to 92 base pairs (bp) on the 3' end, quality filtered and demultiplex them according to the 6 bp unique barcodes. Reads with Phred scores of < 33 were discarded. The barcodes and restriction site residues (6 bp) were removed from the 5' end, and this resulted in a final sequence length of 80 bp.
RAD analysis The software program STACKS version 2.2 (Catchen et al. 2011, 2013) was used to identify orthologous sequences among individuals. In order to optimize the STACKS protocol, we followed published guidelines (Rochette and Catchen 2017). Briefly, we first identified 14 individuals with the highest sequencing coverage and created a catalog of loci based on these samples. (using the ustacks and cstacks components of STACKS). In addition, we optimized the values of the parameters M and n (M and n values were kept identical to each other), by varying them from 1 to 9 as recommended. We found that the optimal value for M and n was 3 for Siganus rivulatus, and 4 for Stephanolepis diaspros. We then included the remaining samples using ustacks.
Analysis of population structure Population structure was determined using all samples for Siganus rivulatus and two separate datasets for Stephanolepis diaspros. The first dataset only included samples adjacent to the Suez Canal, as for better comparison with S. rivulatus. A second dataset included also samples from Djerba, Tunisia, to assess large spatial scales in that species. Using the population component in STACKS, we further filtered the dataset by retaining loci which aligned in ≥ 80% of individuals (r command, − r 0.8) in every population. In order to remove paralogs, we used the minor alleles function of STACKS following recommendations (–min_mav 0.05). We then generated genepop files using the populations component in STACKS with the write_single_snp option (a single SNP is kept for each locus), which were converted afterwards using the program PGDSpider V.2.1.0.3 (Lischer and Excoffier 2012).
Population structure was analyzed using common population genetic metrics estimated by GENODIVE (Fst, F’st, and heterozygosities), and STACKS (Fst), as well as using STRUCTURE, PCA, and DAPC approaches. Structure files from the STACKS populations output were analyzed using a Bayesian approach in STRUCTURE version 2.3.4 (Pritchard et al. 2000). Ten replicate runs were performed for a range of K from one to seven, with 10,000 iterations as the burn-in parameter and 100,000 iterations under the admixture model. The highest likelihood for K was estimated according to the Evanno method (Evanno et al. 2005) implemented in Structure Harvester (Earl and VonHoldt 2012). We performed a Principal Component Analysis (PCA) when analyzing two populations, and when more than two populations were analyzed, we used a Discriminant Analysis of Principal Components (DAPC), (Jombart et al. 2010; Miller et al. 2020), which combines the benefits of discriminant and principal component analyses, and is particularly useful to study differences between clusters (i.e., sites or populations) as it utilizes a multivariate approach to explore the entire variation in the data while minimizing that within clusters. These analyses were performed using the ADEGENET package in R (Jombart 2008; R CoreTeam 2016) using the vcf file produced by the STACKS populations output used as an input file. The algorithm find.clusters identified the plausible number of clusters by comparing Bayesian Information Criterion (BIC) values, and the cross-validation tool xvalDapc determined the number of principal components that were retained.
Outlier loci analyses We examined genetic structure in neutral and outlier loci, separately, to investigate the possibility that different factors have shaped population divergence in unique patterns. Outlier loci were identified using PCADAPT implemented in R (Privé et al. 2020). We used the STACKS populations output to determine Fst (Phi st) values and their distributions to separate loci under directional (Loci with large Fst, i.e. Fst > 0.1), and stabilizing selection. Loci identifiers were then placed in “whitefiles”, which were used to run population scripts again. Although working with outliers might incorporate a series of shortfalls (Bierne et al. 2011, 2013; Lotterhos and Whitlock 2015), they are commonly used to show evidence of the diverging effects of selection between populations (Gaither et al. 2015; Longo and Bernardi 2015; Bernardi et al. 2016; Stockwell et al. 2016).
Functional analyses
All outlier loci were compared to GenBank entries with BLAST, where E-values of 0.001 and below were kept and recorded (probability of obtaining the same result by chance < 0.001). When protein coding matching sequences were found, they were classified using KEGG assignments (Ogata et al. 1999; Kanehisa et al. 2008).
Results
Loci and polymorphism statistics
Genomic DNA was sequenced from a total of 78 samples (38 Siganus rivulatus and 40 Stephanolepis diaspros) in an Illumina lane producing approximately 100 million reads. After passing all filters in the population scripts, STACKS identified a total of 31,249 and 17,831 loci for S. rivulatus and S. diaspros, respectively (Table 2). When grouping individuals in only two populations (Red Sea and Mediterranean), STACKS identified 27,836 and 12,344 loci, respectively (Table 2).
Observed heterozygosities were similar in the two species, 0.187 and 0.196 for S. rivulatus and S. diaspros, respectively (Table 2). A decrease in observed heterozygosity in invading Mediterranean populations was not observed in either species. Indeed, those values were very similar in Red Sea and Mediterranean populations for both species: 0.161–0.187 for Red Sea and Mediterranean populations of S. rivulatus, 0.185 and 0.179 for Red Sea and Mediterranean populations of S. diaspros.
Population structure
All loci
Classical measures of population structure showed that structure was very weak between Red Sea and Mediterranean populations of S. rivulatus: Fst = 0.015, Fst = 0.005, F’st = 0.006 (Table 2). The locus with the highest Fst had a value of 0.506. In contrast, S. diaspros showed higher values, suggesting population structure between Red Sea and Mediterranean populations: Fst = 0.042, Fst = 0.058, F’st = 0.069 (Table 2). The locus with the highest Fst showed a higher value of 0.687.
A principal component analysis (PCA) mirrored the results described above. Red Sea and Mediterranean populations did not separate for S. rivulatus, while they showed structure in S. diaspros (Fig. 2). A STRUCTURE analysis, again, was consistent with both PCA and population structure metrics, showing no structure in S. rivulatus and distinct patterns between Red Sea and Mediterranean populations of S. diaspros (Fig. 3).
Discriminant analysis of principal components (DAPC) cluster plots of Mediterranean and Red Sea populations (p = 2) of Siganus rivulatus and Stephanolepis diaspros based on 27,836 and 12,344 loci, respectively (upper panel). Lower panel shows a smilar analysis based on outlier loci (15 and 22 loci, respectively). Plots were created in R using the adegenet package
STRUCTURE plots of Siganus rivulatus and Stephanolepis diaspros individuals from the Red Sea and the Mediterranean based on neutral and outlier SNPs. STRUCTURE plots (K = 2) with Bayesian assignment of individuals (vertical bars) based on presumed neutral loci (upper panel) and outlier loci (loci suspected to be under selection (lower panel)
When considering each population independently (as opposed to separating individuals in only Red Sea and Mediterranean populations), results were consistent with what was presented above (Table 2, Supplementary Fig. 1).
Outlier loci and genes under selection
A combination of PCADAPT and STACKS identified loci under potential directional selection (outlier loci). The number of outlier loci was lower in S. rivulatus than in S. diaspros: 15 versus 22 for the Red Sea and Mediterranean comparison, 74 versus 201 when considering all populations (Table 2). As expected, values of Fst were much higher when considering outlier loci, and were again lower in S. rivulatus than in S. diaspros (0.154 and 0.22, respectively, Table 2). Patterns observed with PCAs and STRUCTURE were consistent with the patterns based on all loci as described above, but the separation of populations was even more pronounced in S. diaspros, while still not evident in S. rivulatus (Figs. 2 and 3).
Outlier loci were compared to GenBank sequences. Protein coding genes were identified for six and five loci for S. rivulatus and S. diaspros, respectively (Table 3). The high proportion of protein coding genes within the outlier loci (40% and 22.7% in S. rivulatus and S. diaspros) is consistent with the assumption that outlier loci are indicative of selection. Three genes were identified as ionic transporters (Table 3), which may play an important role in osmoregulation in fishes.
Additional population
Considering the structure obtained in S. diaspros, we sampled an additional population further away from the Suez Canal in the Mediterranean (Djerba, Tunisia). The addition of this population gave us insight into the pattern of population structure in that species. Patterns of population structure described above were even more pronounced with the addition of this population, with higher levels of population structure (Fst = 0.069, F’st = 0.081, Table 2), more divergent populations as analyzed by PCA and DAPC (Supplementary Fig. 1), and more outlier loci (332 loci, Table 2). We analyzed the composition of the protein coding genes associated with the outlier loci of this analysis, and found that the largest family of genes under selection were related with Environmental Processing Information, comprising 26% of all the loci under selection (Fig. 4), suggesting potential for local adaptation to a new environment.
Discussion
Successful biological invasions generally share common attributes, such as large numbers of invading individuals that carry high genetic diversity (Blackburn et al. 2015), genomic regions under selection (Bernardi et al. 2016; Stuart et al. 2020; Barbosa 2021), or hybridization strategies (Blackwell et al. 2020; Valencia-Montoya et al. 2020; Popovic and Bernatchez 2021). In the case of Lessepsian bionvasions, most fish populations show an absence of genetic bottleneck, and a lack of population structure between Red Sea and Mediterranean populations (Bernardi et al. 2010; Chiesa et al. 2019). Thus far, however, little information has been available about one important aspect of the invasion, namely the genetics of populations around their entry gate, ie. in the immediate vicinity of the Suez Canal. The goal of this study was to fill this knowledge gap by looking, as a case study, at the genomics of the rabbitfish Siganus rivulatus and the filefish Stephanolepis diaspros.
Importantly, the focal region of this study is very limited spatially, with the Canal being a little less than 200 km in length, thus apparently not conducive to strong genetic structure in marine organisms (White et al. 2010; Selkoe and Toonen 2011; Pinsky et al. 2017). This expectation (no genetic differentiation) is consistent with some of our results. Indeed, the PCA and STRUCTURE analyses, as well as the classic metrics for population structure and genetic diversity, all indicated that the rabbitfish, S. rivulatus, showed little genetic structure between Red Sea and Mediterranean populations. In contrast, and surprisingly, the filefish, S. diaspros, showed strong population structure, even at such small spatial scale. This population structure mirrored the pattern seen at larger spatial scales when we added a further population from Djerba, Tunisia. This pattern is consistent with the prediction that strong population structure at larger scales is a good proxy for population structure in invading populations (Gaither et al. 2013; Jackson et al. 2015). Life-history traits are known to influence connectivity of marine fish populations (Chopelet et al. 2009). In this regard, the monogamous habit of filefishes and their complex mating system (Rim and Bradai 2011) may explain the genetic patterns that we observed. Yet, the strong population structure across the Suez Canal that was observed in S. diaspros is worth exploring in light of the main determinants, dispersal mechanisms v.s. selection, that possibly affect the genetic structure of these invaders.
Dispersal mechanisms
Crucial gaps in knowledge on dispersal mechanisms still hinder a thorough understanding of patterns of genetic diversity of Lessepsian fishes, but some hypotheses may be proposed. In marine fish populations, where the pelagic larval stage may last weeks to months, dispersal capability is often equated to gene flow and genetic divergence (Slatkin 1987; Selkoe and Toonen 2011; Medina et al. 2018). Due to the difficulties of tracking the fate of propagules, no direct observation exists on the pattern of dispersal through the 200 km long Suez Canal. Oceanographic models for the Mediterranean provide realistic pathways for potential dispersal of fish propagules (Bouzaiene et al. 2020). Within the Suez Canal, water movement models show that, while some recent southward migration may be possible, the main water transport is northward, from the Red Sea into the Mediterranean, as expected from the min observed biological invasions (Biton 2020). While it is assumed that most invasions occur via passive larval transport, parasitological evidence supports that crossing is achieved, at least in part, by adult individuals that actively swim from the Red Sea into the Mediterranean (Diamant 2010). In this respect, the limited swimming ability of adult filefishes, and their territoriality (Kawase and Nakazono 1996), could explain the observed genetic differentiation between Red Sea and Mediterranean populations, resulting from a reduced number of propagules entering the Mediterranean. The idea that dispersal characteristics may have influenced the difference in population structure in S. rivulatus and S. diaspros is also consistent with previous studies that have shown some evidence of structure in Stephanolepis filefishes (Yoon et al. 2012), and a lack of structure in Mediterranean rabbitfishes (Bonhomme et al. 2003; Hassan et al. 2003; Hassan and Bonhomme 2005; Azzurro et al. 2006). It is important to note, however, that other species of Pacific rabbitfishes, do show some, albeit low, levels of population structure in Indonesia and in Guam (Priest et al. 2012; Bramandito et al. 2018).
Natural selection and preadaptations
The environmental conditions encountered by Lessepsian migrants during their transition through the Suez Canal are very diverse: the Red Sea tends to be warmer and saltier than the Mediterranean, and although the salinity of the bitter lakes is variable, it is generally still much saltier than either Red Sea or Mediterranean (El-Bassat 2008). Our results reinforce the idea that the ability of invaders to cope with osmotic changes, in addition to simple dispersal capabilities, is an important factor.
Dispersal (and its genetic consequence, gene flow) counter the potential for local adaptation. However, when selection is strong, it may overcome the homogenizing effects of gene flow, to the point of creating population structure and even occasionally resulting in speciation (Nosil 2008; Savolainen et al. 2013; Arick et al. 2020). Considering the strong environmental gradient encountered by Lessepsian migrants in the Suez Canal, genes under selection involved with osmoregulation have been found in the bluespotted cornetfish, a very successful Lessepsian migrant (Bernardi et al. 2016). Therefore, we looked for evidence of selection in the face of gene flow in the two focal species of this study. In this study, both focal species showed loci under directional selection, and for both species, genes under selection included functions that are potentially involved with osmoregulation. Interestingly, selection was uncovered in the face of low (S. diaspros) and high (S. rivulatus) gene flow, indicating that selection must be very strong to be detectable.
Conclusion
With the advent of genomic approaches, the role of genes under selection has been pushed to the forefront of the field. In general, invading individuals have been present in their new environment for too short a time for mutations to accumulate, and selection to act upon them. A more likely scenario is that variability is present in the original population, and either a large number of individuals invades, and only locally adapted individuals survive, or alternatively, only preadapted individuals are capable of thriving in their new environment. In order to distinguish those scenarios, one must look at the very early phase of invasion. Results obtained in this study suggest that individuals showing signatures of selection are present in the vicinity of Suez Canal, suggesting that at least some Lessepsian invaders show preadapation characteristics right out of the gate.
Data availability
All fastq sequence files are available from GenBank at the National Center for Biotechnology Information short-read archive (NCBI-SRA) database (accession numbers: SAMN22938593- SAMN22938670, under project: PRJNA778315). Associated metadata are also available at GeOMe (https://n2t.net/ark:/21547/Dxl2).
References
Arick LN, Dimeo M, Rautsaw RM et al (2020) Genomic adaptations to salinity resist gene flow in the evolution of Floridian watersnakes. Mol Biol Evol 38:745–760. https://doi.org/10.1093/molbev/msaa266
Azzurro E, Golani D, Bucciarelli G, Bernardi G (2006) Genetics of the early stages of invasion of the Lessepsian rabbitfish Siganus luridus. J Exp Mar Bio Ecol 333:190–201. https://doi.org/10.1016/j.jembe.2005.12.002
Azzurro E, Goren M, Diamant A et al (2015) Establishing the identity and assessing the dynamics of invasion in the Mediterranean Sea by the dusky sweeper, Pempheris rhomboidea Kossmann & Räuber, 1877 (Pempheridae, Perciformes). Biol Invasions 17:815–826. https://doi.org/10.1007/s10530-014-0836-5
Baird NA, Etter PD, Atwood TS et al (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3:e3376. https://doi.org/10.1371/journal.pone.0003376
Baker HG, Stebbins GL (1965) The genetics of colonizing species. Academic Press, New York, NY
Barbosa S (2021) Is a handful of genes responsible for the common starling invasion success ? Mol Ecol 30:1361–1363. https://doi.org/10.1111/mec.15794
Bariche M, Bernardi G (2009) Lack of a genetic bottleneck in a recent Lessepsian bioinvader, the blue-barred parrotfish, Scarus ghobban. Mol Phylogenet Evol 53:592–595. https://doi.org/10.1016/j.ympev.2009.06.017
Bariche M, Letourneur Y, Harmelin-Vivien M (2004) Temporal fluctuations and settlement patterns of native and Lessepsian herbivorous fishes on the Lebanese coast (eastern Mediterranean). Environ Biol Fishes 70:81–90
Bariche M, Sadek R, Azzurro E (2009) Fecundity and condition of successful invaders: Siganus rivulatus and S. luridus (Actinopterygii: Perciformes: Siganidae) in the eastern Mediterranean Sea. Acta Ichthyol Piscat 39:11–18. https://doi.org/10.3750/AIP2009.39.1.03
Bernardi G, Azzurro E, Golani D, Miller MR (2016) Genomic signatures of rapid adaptive evolution in the bluespotted cornetfish, a Mediterranean Lessepsian invader. Mol Ecol 25:3384–3396. https://doi.org/10.1111/mec.13682
Bernardi G, Golani D, Azzurro E (2010) The genetics of Lessepsian bioinvasions. In: Golani D, Applebaum-Golani B (eds) Fish invasions of the Mediterranean Sea: change and renewal. Pensoft Publishers, Sofia-Moscow, pp 71–84
Betancur RR, Hines A, Arturo AP et al (2011) Reconstructing the lionfish invasion: insights into greater Caribbean biogeography. J Biogeogr 38:1281–1293. https://doi.org/10.1111/j.1365-2699.2011.02496.x
Bierne N, Roze D, Welch J (2013) Pervasive selection or is it … ? why are Fst outliers sometimes so frequent ? Mol Ecol 22:2061–2064
Bierne N, Welch J, Loire E et al (2011) The coupling hypothesis: why genome scans may fail to map local adaptation genes. Mol Ecol 20:2044–2072. https://doi.org/10.1111/j.1365-294X.2011.05080.x
Biton E (2020) Possible implications of sea level changes for species migration through the Suez Canal. Sci Rep 10:21195. https://doi.org/10.1038/s41598-020-78313-2
Blackburn TM, Lockwood JL, Cassey P (2017) The influence of numbers on invasion success. In: Barrett S, Colautti R, Dlugosch KM, Rieseberg LH (eds) Invasion genetics: the baker and stebbins legacy. John Wiley and Sons, pp 25–39
Blackburn TM, Lockwood JL, Cassey P (2015) The influence of numbers on invasion success. Mol Ecol. https://doi.org/10.1111/mec.13075
Blackwell T, Ford AGP, Ciezarek AG et al (2020) Newly discovered cichlid fish biodiversity threatened by hybridization with non-native species. Mol Ecol. https://doi.org/10.1111/mec.15638
Bonhomme F, Baranes AVI, Golani D, Harmelin-Vivien M (2003) Lack of mitochondrial differentiation between Red Sea and Mediterranean populations of the Lessepsian rabbitfish, Siganus rivulatus (Perciformes: Siganidae)*. Sci Mar 67:215–217
Bos AR, Ogwang J, Bariche M et al (2020) Anti-Lessepsian migration rectified: the Comber Serranus cabrilla (L. 1758) existed in the red sea prior to the Suez Canal opening. Mar Biol 167:1–10. https://doi.org/10.1007/s00227-020-03748-0
Bouzaiene M, Menna M, Poulain PM et al (2020) Analysis of the surface dispersion in the Mediterranean sub-basins. Front Mar Sci 7:1–20. https://doi.org/10.3389/fmars.2020.00486
Bramandito A, Subhan B, Prartono T, Anggraini NP (2018) Genetic diversity and population structure of Siganus fuscescens across urban reefs of Seribu Islands, Northern of Jakarta, Indonesia. Biodiversitas 19:1993–2002. https://doi.org/10.13057/biodiv/d190603
Bryan PG, Madraisau BB (1977) Larval rearing and development of Siganus lineatus (Pisces: Siganidae) from hatching through metamorphosis. Aquaculture 10:243–252
Bucciarelli G, Golani D, Bernardi G (2002) Genetic cryptic species as biological invaders: the case of a Lessepsian fish migrant, the hardyhead silverside Atherinomorus lacunosus. J Exp Mar Bio Ecol 273:143–149
Catchen J, Hohenlohe PA, Bassham S et al (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140. https://doi.org/10.1111/mec.12354
Catchen JM, Amores A, Hohenlohe P et al (2011) Stacks: building and genotyping loci de novo from short-read sequences. G3 Bethesda 1:171–182. https://doi.org/10.1534/g3.111.000240
Chiesa S, Azzurro E, Bernardi G (2019) The genetics and genomics of marine fish invasions: a global review. Rev Fish Biol Fish 29:837–859. https://doi.org/10.1007/s11160-019-09586-8
Chopelet J, Waples RS, Mariani S (2009) Sex change and the genetic structure of marine fish populations. Fish Fish. https://doi.org/10.1111/j.1467-2979.2009.00329.x
Deidun A, Castriota L, Falautano M (2015) Documenting the occurrence of the Lessepsian fish Stephanolepis diaspros within the strait of sicily, central Mediterranean. J Black Sea/Mediterranean Environ 21:1–11
Diamant A (2010) Red-med immigration: a fish parasitology perspective, with special reference to the Myxosporea. In: Golani D, Appelbaum-Golani B (eds) Fish invasions of the Mediterranean Sea: change and renewal. Pensoft Publishers, Sofia-Moscow, pp 85–97
Dlugosch KM, Parker IM (2008) Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol Ecol 17:431–449. https://doi.org/10.1111/j.1365-294X.2007.03538.x
Earl DA, VonHoldt BM (2012) Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4:359–361
El-Bassat RA (2008) Composition and abundance of the zooplankton community in the bitter lakes, Egypt, in relation to environmental factors. Afr J Aquat Sci 33:233–240. https://doi.org/10.2989/AJAS.2008.33.3.5.617
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software Structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Gaither MR, Bernal MA, Coleman RR et al (2015) Genomic signatures of geographic isolation and natural selection in coral reef fishes. Mol Ecol 24:1543–1557. https://doi.org/10.1111/mec.13129
Gaither MR, Bowen BW, Toonen RJ (2013) Population structure in the native range predicts the spread of introduced marine species. Proc R Soc B Biol Sci London 280:20130409
Golani D (2010) Colonization of the Mediterranean by red sea fishes via the Suez Canal: Lessepsian migration. In: Golani D, Appelbaum-Golani B (eds) Fish invasions of the Mediterranean Sea: change and renewal. Pensoft Publishers, Sofia, pp 145–188
Golani D, Azzurro E, Corsini-Foka M et al (2007) Genetic bottlenecks and successful biological invasions: the case of a recent Lessepsian migrant. Biol Lett 3:541–545. https://doi.org/10.1098/rsbl.2007.0308
Golani D, Ritte U (1999) Genetic relationship in goatfishes (Mullidae : Perciformes) of the Red Sea and the Mediterranean, with remarks on Suez Canal migrants*. Sci Mar 63:129–135
Hassan M, Bonhomme F (2005) No reduction in neutral variability of mitochondrial and nuclear genes for a Lessepsian migrant, Upeneus moluccensis. J Fish Biol 66:865–870. https://doi.org/10.1111/j.1095-8649.2005.00643.x
Hassan M, Harmelin-Vivien M, Bonhomme F (2003) Lessepsian invasion without bottleneck: example of two rabbitfish species (Siganus rivulatus and Siganus luridus). J Exp Mar Bio Ecol 291:219–232. https://doi.org/10.1016/S0022-0981(03)00139-4
Holland BS (2001) Invasion without a bottleneck: Microsatellite variation in natural and invasive populations of the brown Mussel Perna perna (L). Mar Biotechnol 3:407–415. https://doi.org/10.1007/s1012601-0060-Z
Jackson AM, Tenggardjaja K, Perez G et al (2015) Phylogeography of the bluespotted cornetfish, Fistularia commersonii: a predictor to bioinvasion success? Mar Ecol 36:887–896
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. https://doi.org/10.1371/journal.pcbi.1000455
Kanehisa M, Araki M, Goto S et al (2008) KEGG for linking genomes to life and the environment. Nucl Acids Res 36:480–484. https://doi.org/10.1093/nar/gkm882
Katsanevakis S, Poursanidis D, Hoffman R et al (2020) Unpublished Mediterranean records of marine alien and cryptogenic species. BioInvasions Rec 9:165–182
Katsanevakis S, Tempera F, Teixeira H (2016) Mapping the impact of alien species on marine ecosystems: the Mediterranean Sea case study. Divers Distrib 22:694–707. https://doi.org/10.1111/ddi.12429
Kawase H, Nakazono A (1996) Two alternative female tactics in the polygynous mating system of the threadsail filefish, Stephanolepis cirrhifer (Monacanthidae). Ichthyol Res 43:315–323
Lam TJ (1974) Siganids: their biology and mariculture potential. Aquaculture 3(4):325–354
Lischer HEL, Excoffier L (2012) PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28:298–299. https://doi.org/10.1093/bioinformatics/btr642
Longo G, Bernardi G (2015) The evolutionary history of the embiotocid surfperch radiation based on genome-wide RAD sequence data. Mol Phylogenet Evol 88:55–63. https://doi.org/10.1016/j.ympev.2015.03.027
Lotterhos KE, Whitlock MC (2015) The relative power of genome scans to detect local adaptation depends on sampling design and statistical method. Mol Ecol 24:1031–1046. https://doi.org/10.1111/mec.13100
Mancera-Rodríguez NJ, Castro-Hernández JJ (2015) Reproductive biology of the planehead filefish Stephanolepis hispidus (Pisces: Monacanthidae), in the Canary Islands area. Ichthyol Res 62:258–267. https://doi.org/10.1007/s10228-014-0435-6
Medina I, Cooke GM, Ord TJ (2018) Walk, swim or fly ? Locomotor mode predicts genetic differentiation in vertebrates. Ecol Lett 21:638–645. https://doi.org/10.1111/ele.12930
Miller JM, Cullingham CI, Peery RM (2020) The influence of a priori grouping on inference of genetic clusters: simulation study and literature review of the DAPC method. Heredity (edinb). https://doi.org/10.1038/s41437-020-0348-2
Miller MR, Brunelli JP, Wheeler PA et al (2012) A conserved haplotype controls parallel adaptation in geographically distant salmonid populations. Mol Ecol 21:237–249. https://doi.org/10.1111/j.1365-294X.2011.05305.x
Miller MR, Dunham JP, Amores A et al (2007) Rapid and cost-effective polymorphism identification and genotyping using restriction site associated DNA (RAD) markers. Genome Res 17:240–248. https://doi.org/10.1101/gr.5681207
Nosil P (2008) Speciation with gene flow could be common. Mol Ecol 17:2103–2106. https://doi.org/10.1186/1742-9994-4-11.Schmitt
Ogata H, Goto S, Sato K et al (1999) KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res 27:29–34. https://doi.org/10.1093/nar/27.1.29
Pallaoro A, Dulc J (2004) First record of the marbled spinefoot Siganus rivulatus (Pisces: Siganidae) in the Adriatic Sea. J Mar Biol Assoc UK 84:1087–1088
Pinsky ML, Saenz-Agudelo P, Salles OC et al (2017) Marine dispersal scales are congruent over evolutionary and ecological time. Curr Biol 27:149–154. https://doi.org/10.1016/j.cub.2016.10.053
Popovic I, Bernatchez L (2021) Uncovering endemism in a lake of invasive species introgression. Mol Ecol. https://doi.org/10.1111/mec.15801
Popper D, Pitt R, Zohar Y (1979) Experiments on the propagation of red sea siganids and some notes on their reproduction in nature. Aquaculture 16:177–181
Por FD (1978) Lessepsian migration: the influx of red sea biota into the Mediterranean by way of the Suez Canal. Springer-Verlag, Berlin
Priest MA, Halford AR, McIlwain JL (2012) Evidence of stable genetic structure across a remote island archipelago through self-recruitment in a widely dispersed coral reef fish. Ecol Evol 2:3195–213. https://doi.org/10.1002/ece3.260
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Privé F, Luu K, Vilhjálmsson BJ et al (2020) Performing highly efficient genome scans for local adaptation with R package pcadapt version 4. Mol Biol Evol 37:2153–2154. https://doi.org/10.1093/molbev/msaa053
R CoreTeam R (2016) R: a language and environment for statistical computing
Rim Z-K, Bradai M-N (2011) Reproductive biology of the lessepsian reticulated leatherjacket Stephanolepis diaspros (Fraser: Brunner, 1940) in the gulf of gabes (Eastern Mediterranean Sea). Rev Fish Biol Fish 21:641–648. https://doi.org/10.1007/s11160-011-9202-1
Rius M, Turon X, Bernardi G et al (2015) Marine invasion genetics: from spatio-temporal patterns to evolutionary outcomes. Biol Invasions 17:869–885. https://doi.org/10.1007/s10530-014-0792-0
Rochette NC, Catchen JM (2017) Deriving genotypes from RAD-seq short-read data using stacks. Nat Protoc 12:2640–2659. https://doi.org/10.1038/nprot.2017.123
Rogers JS, Hare JA, Lindquist DG (2001) Otolith record of age, growth, and ontogeny in larval and pelagic juvenile Stephanolepis hispidus (Pisces: Monacanthidae). Mar Biol 138:945–953. https://doi.org/10.1007/s002270000521
Roman J, Darling JA (2007) Paradox lost: genetic diversity and the success of aquatic invasions. Trends Ecol Evol 22:454–464. https://doi.org/10.1016/j.tree.2007.07.002
Savolainen O, Lascoux M, Merilä J (2013) Ecological genomics of local adaptation. Nat Rev Genet 14:807–820. https://doi.org/10.1038/nrg3522
Sax DF, Stachowicz JJ, Gaines SD (2005) Species invasions: insights into ecology, evolution and biogeography. Sinauer Associates Incorporated
Sax DF, Stachowicz JJ, Brown JH et al (2007) Ecological and evolutionary insights from species invasions. Trends Ecol Evol 22:465–471
Selkoe K, Toonen R (2011) Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar Ecol Prog Ser 436:291–305. https://doi.org/10.3354/meps09238
Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science (80-) 236:787–792. https://doi.org/10.1126/science.3576198
Stamouli C, Akel EHKH, Azzurro E et al (2017) New Mediterranean biodiversity records (December 2017). Mediterr Mar Sci 18:534–556
Steinitz W (1927) Beiträge zur Kenntnis der Küstenfauna Palästinas. I. Pubbl Della Stn Zool Di Napoli 8:311–353
Stockwell BL, Larson WA, Waples RK et al (2016) The application of genomics to inform conservation of a functionally important reef fish (Scarus niger) in the Philippines. Conserv Genet 17:239–249. https://doi.org/10.1007/s10592-015-0776-3
Stuart KC, Cardilini APA, Cassey P et al (2020) Signatures of selection in a recent invasion reveal adaptive divergence in a highly vagile invasive species. Mol Ecol. https://doi.org/10.1111/mec.15601.10.1111/mec.15601
Stuart KC, Cardilini APA, Cassey P et al (2021) Signatures of selection in a recent invasion reveal adaptive divergence in a highly vagile invasive species. Mol Ecol 30:1419–1434. https://doi.org/10.1111/mec.15601
Tenggardjaja K, Jackson AM, Leon F et al (2014) Genetics of a Lessepsian sprinter the bluespotted cornetfish, Fistularia commersonii. Isr J Ecol Evol 59:181–185
Tiralongo F, Lillo AO, Tibullo D et al (2019) Monitoring uncommon and non-indigenous fishes in Italian waters: One year of results for the AlienFish project. Reg Stud Mar Sci 28:100606
Tortonese E (1984) Mediterranean fishes present in the red sea: pan-oceanic and anti-Lessepsian species. Cybium 8:99–102
Valencia-Montoya WA, Elfekih S, North HL et al (2020) Adaptive introgression across semipermeable species boundaries between local Helicoverpa zea and invasive Helicoverpa armigera moths. Mol Biol Evol 37:2568–2583. https://doi.org/10.1093/molbev/msaa108
White C, Selkoe KA, Watson J et al (2010) Ocean currents help explain population genetic structure. Proc Biol Sci 277:1685–94. https://doi.org/10.1098/rspb.2009.2214
Xue D-X, Carranza A, Graves J et al (2018) Successful worldwide invasion of the veined rapa whelk, Rapana venosa, despite a dramatic genetic bottleneck. Biol Invasions 20:3297–3314. https://doi.org/10.1007/s10530-018-1774-4
Yoon M, Park W, Nam YK, Kim DS (2012) Shallow population genetic structures of Thread-sail Filefish (Stephanolepis cirrhifer) populations from Korean coastal waters. Asian-Australas J Anim Sci 25:170–176
Zenetos A, Galanidi M (2020) Mediterranean non indigenous species at the start of the 2020s: recent changes. Mar Biodivers Rec 13(10):1–17
Zenetos Α, Gofas S, Morri C et al (2012) Alien species in the Mediterranean Sea by 2012. A contribution to the application of European Union’s marine strategy framework directive (MSFD). Part 2. Introduction trends and pathways. Mediterr Mar Sci 13:328–352
Acknowledgements
Funding for this work was provided by the Committee on Research, COR-UCSC, to GB.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10530_2021_2704_MOESM1_ESM.pdf
Discriminant analysis of principal components (DAPC) cluster plots of all populations of Siganus rivulatus and Stephanolepis diaspros based on 27,836 and 12,344 loci, respectively (upper panel). Lower panel shows a smilar analysis based on outlier loci (15 and 22 loci, respectively). Plots were created in R using the adegenet package. (PDF 227 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Azzurro, E., Nourigat, M., Cohn, F. et al. Right out of the gate: the genomics of Lessepsian invaders in the vicinity of the Suez Canal. Biol Invasions 24, 1117–1130 (2022). https://doi.org/10.1007/s10530-021-02704-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-021-02704-3