Abstract
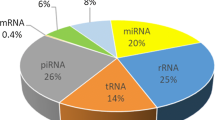
Mammalian spermatozoa comprises both coding and non-coding RNAs, which are traditionally believed to be a residual of spermatogenesis. The differential expression level of spermatozoal RNAs is also observed between fertile and infertile human, thereby anticipated as potential molecular marker of male fertility. This study investigated the transcriptome profile of goat (Capra hircus) spermatozoa. The sperm transcriptome was analyzed by three different methods viz. RLM-RACE, long-read RNA sequencing (RNAseq) in Nanopore™ platform, and short-read RNAseq in Illumina™ platform. The Illumina™ sequencing discovered 16,604 transcripts with 357 mRNAs having FPKM (fragments per kilobase per million mapped reads) of more than five. The spermatozoal RNA suite included mRNA (94%), rRNA (3%), miscRNA (1%), circRNA (1%), miRNA (1%), etc. This study also predicted circRNAs (127), lncRNAs (655), and imprinted genes (160) that have potential role in male reproduction. The gene ontology analysis revealed the involvement of spermatozoal RNA in regulating male meiosis (TET3, STAT5B), capacitation (ACRBP, CATSPER4), sperm motility (GAS8, TEKT2), spermatogenesis (ADAMTS2, CREB3L4), etc. The spermatozoal RNA were also associated with different biological pathways viz. Wnt signaling pathway, cAMP signaling pathway, AMPK signaling pathway, and MAPK signaling pathways having potential role in spermatogenesis. Overall, this study enlightened the suite of spRNA transcripts in goat and their relevance in male fertility for diagnostic approach.








Similar content being viewed by others
References
Alvarez-Rodriguez M, Martinez C, Wright D et al (2020) The transcriptome of pig spermatozoa, and its role in fertility. Int J Mol Sci 21:1572. https://doi.org/10.3390/ijms21051572
Babakhanzadeh E, Khodadadian A, Rostami S et al (2020) Testicular expression of TDRD1, TDRD5, TDRD9 and TDRD12 in azoospermia. BMC Med Genet 21:1–7. https://doi.org/10.1186/s12881-020-0970-0
Cannarella R, Condorelli RA, La VS et al (2019) IGF2 and IGF1R mRNAs are detectable in human spermatozoa. World J Men’s Health 37:24–30. https://doi.org/10.5534/WJMH.190070
Cannarella R, Condorelli RA, Mongioì LM et al (2020) Molecular biology of spermatogenesis: novel targets of apparently idiopathic male infertility. Int J Mol Sci. https://doi.org/10.3390/ijms21051728
Capra E, Turri F, Lazzari B et al (2017) Small RNA sequencing of cryopreserved semen from single bull revealed altered miRNAs and piRNAs expression between high- and low-motile sperm populations. BMC Genomics 18:1–13. https://doi.org/10.1186/s12864-016-3394-7
Card CJ, Anderson EJ, Zamberlan S et al (2013) Cryopreserved bovine spermatozoal transcript profile as revealed by high-throughput ribonucleic acid sequencing1. Biol Reprod 88:1–9. https://doi.org/10.1095/biolreprod.112.103788
Castillo J, Jodar M, Oliva R (2018) The contribution of human sperm proteins to the development and epigenome of the preimplantation embryo. Hum Reprod Update 24:535–555. https://doi.org/10.1093/humupd/dmy017
Chalmel F, Lardenois A, Evrard B et al (2012) Global human tissue profiling and protein network analysis reveals distinct levels of transcriptional germline-specificity and identifies target genes for male infertility. Hum Reprod 27:3233–3248. https://doi.org/10.1093/humrep/des301
Chen J, Gu Y, Zhang Z et al (2016) Deficiency of SPATA46, a novel nuclear membrane protein, causes subfertility in male mice. Biol Reprod 95:58. https://doi.org/10.1095/biolreprod.116.140996
Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. https://doi.org/10.1093/bioinformatics/bty560
Chu C, Yu L, Wu B et al (2017) A sequence of 28S rRNA-derived small RNAs is enriched in mature sperm and various somatic tissues and possibly associates with inflammation. J Mol Cell Biol 9:256–259. https://doi.org/10.1093/jmcb/mjx016
Corral-Vazquez C, Salas-Huetos A, Blanco J et al (2019) Sperm microRNA pairs: new perspectives in the search for male fertility biomarkers. Fertil Steril 112:831–841. https://doi.org/10.1016/j.fertnstert.2019.07.006
Cullinane DL, Chowdhury TA, Kleene KC (2015) Mechanisms of translational repression of the Smcp mRNA in round spermatids. Reproduction 149:43–54. https://doi.org/10.1530/REP-14-0394
Das PJ, McCarthy F, Vishnoi M et al (2013) Stallion sperm transcriptome comprises functionally coherent coding and regulatory RNAs as revealed by microarray analysis and RNA-seq. PLoS ONE 8:1–17. https://doi.org/10.1371/journal.pone.0056535
de Lima AO, Afonso J, Edson J et al (2021) Network analyses predict small RNAs that might modulate gene expression in the testis and epididymis of Bos indicus bulls. Front Genet 12:1–14. https://doi.org/10.3389/fgene.2021.610116
Fagerlind M, Stålhammar H, Olsson B, Klinga-Levan K (2015) Expression of miRNAs in bull spermatozoa correlates with fertility rates. Reprod Domest Anim 50:587–594. https://doi.org/10.1111/rda.12531
Ferlin A, Vinanzi C, Garolla A et al (2006) Male infertility and androgen receptor gene mutations: clinical features and identification of seven novel mutations. Clin Endocrinol (oxf) 65:606–610. https://doi.org/10.1111/j.1365-2265.2006.02635.x
Fu G, Wei Y, Wang X, Yu L (2016) Identification of candidate causal genes and their associated pathogenic mechanisms underlying teratozoospermia based on the spermatozoa transcript profiles. Andrologia 48:576–583. https://doi.org/10.1111/and.12484
Fujihara Y, Satouh Y, Inoue N et al (2012) SPACA1-deficient male mice are infertile with abnormally shaped sperm heads reminiscent of globozoospermia. Development 139:3583–3589. https://doi.org/10.1242/dev.081778
Gao Y, Zhang J, Zhao F (2018) Circular RNA identification based on multiple seed matching. Brief Bioinform 19:803–810. https://doi.org/10.1093/bib/bbx014
Gau BH, Chu IM, Huang MC et al (2008) Transcripts of enriched germ cells responding to heat shock as potential markers for porcine semen quality. Theriogenology 69:758–766. https://doi.org/10.1016/j.theriogenology.2007.11.020
Girault MS, Dupuis S, Ialy-Radio C et al (2021) Deletion of the spata3 gene induces sperm alterations and in vitro hypofertility in mice. Int J Mol Sci 22:1–13. https://doi.org/10.3390/ijms22041959
Gòdia M, Castelló A, Rocco M et al (2020) Identification of circular RNAs in porcine sperm and evaluation of their relation to sperm motility. Sci Rep 10:1–11. https://doi.org/10.1038/s41598-020-64711-z
Grzmil P, Boinska D, Kleene KC et al (2008) Prm3, the fourth gene in the mouse protamine gene cluster, encodes a conserved acidic protein that affects sperm motility. Biol Reprod 78:958–967. https://doi.org/10.1095/biolreprod.107.065706
Harayama H (2013) Roles of intracellular cyclic AMP signal transduction in the capacitation and subsequent hyperactivation of mouse and boar spermatozoa. J Reprod Dev 59:421–430. https://doi.org/10.1262/jrd.2013-056
Jamsai D, O’Connor AE, O’Donnell L et al (2015) Uncoupling of transcription and translation of Fanconi anemia (FANC) complex proteins during spermatogenesis. Spermatogenesis 5:1–9. https://doi.org/10.4161/21565562.2014.979061
Johnson GD, Mackie P, Jodar M et al (2015) Chromatin and extracellular vesicle associated sperm RNAs. Nucleic Acids Res 43:6847–6859. https://doi.org/10.1093/nar/gkv591
Joshi M, Rajender S (2020) Long non-coding RNAs (lncRNAs) in spermatogenesis and male infertility. Reprod Biol Endocrinol 18:103. https://doi.org/10.1186/s12958-020-00660-6
Juma AR, Grommen SVH, O’Bryan MK et al (2017) PLAG1 deficiency impairs spermatogenesis and sperm motility in mice. Sci Rep 7:1–12. https://doi.org/10.1038/s41598-017-05676-4
Kawano M, Kawaji H, Grandjean V et al (2012) Novel small noncoding RNAs in mouse spermatozoa, zygotes and early embryos. PLoS ONE 7:6–13. https://doi.org/10.1371/journal.pone.0044542
Kiani M, Salehi M, Mogheiseh A (2019) MicroRNA expression in infertile men: its alterations and effects. Zygote 27:263–271. https://doi.org/10.1017/S0967199419000340
Koppes E, Shaffer B, Sadovsky E et al (2019) Klf14 is an imprinted transcription factor that regulates placental growth. Placenta 88:61–67. https://doi.org/10.1016/j.placenta.2019.09.013
Lee LK, Foo KY (2014) Recent insights on the significance of transcriptomic and metabolomic analysis of male factor infertility. Clin Biochem 47:973–982. https://doi.org/10.1016/j.clinbiochem.2014.05.053
Li C-Y, Guo Z, Wang Z (2007) TGFbeta receptor saxophone non-autonomously regulates germline proliferation in a Smox/dSmad2-dependent manner in Drosophila testis. Dev Biol 309:70–77. https://doi.org/10.1016/j.ydbio.2007.06.019
Liu T, Cheng W, Gao Y et al (2012) Microarray analysis of microRNA expression patterns in the semen of infertile men with semen abnormalities. Mol Med Rep 6:535–542. https://doi.org/10.3892/mmr.2012.967
Liu XX, Cai L, Liu FJ (2018) An in silico analysis of human sperm genes associated with asthenozoospermia and its implication in male infertility. Medicine 97:1–7. https://doi.org/10.1097/MD.0000000000013338
Malik RK, Lohan IS, Dhanda OP et al (1999) Peritoneal fluid from rabbits or goats as media for in vitro maturation, fertilization and initial culture of caprine oocytes. Anim Reprod Sci 54:195–201. https://doi.org/10.1016/S0378-4320(98)00100-6
Manfrevola F, Chioccarelli T, Cobellis G et al (2020) CircRNA role and circRNA-dependent network (ceRNET) in asthenozoospermia. Front Endocrinol 11:1–14. https://doi.org/10.3389/fendo.2020.00395
Mokánszki A, Molnár Z, Varga Tóthné E et al (2020) Altered microRNAs expression levels of sperm and seminal plasma in patients with infertile ejaculates compared with normozoospermic males. Hum Fertil 23:246–255. https://doi.org/10.1080/14647273.2018.1562241
Navarro-Costa P, Nogueira P, Carvalho M et al (2010) Incorrect DNA methylation of the DAZL promoter CpG island associates with defective human sperm. Hum Reprod 25:2647–2654. https://doi.org/10.1093/humrep/deq200
Ostermeier GC, Miller D, Huntriss JD et al (2004) Reproductive biology: delivering spermatozoan RNA to the oocyte. Nature 429:154. https://doi.org/10.1038/429154a
Özbek M, Hitit M, Kaya A et al (2021) Sperm functional genome associated with bull fertility. Front Vet Sci 8:1–17. https://doi.org/10.3389/fvets.2021.610888
Parthipan S, Selvaraju S, Somashekar L et al (2017) Spermatozoal transcripts expression levels are predictive of semen quality and conception rate in bulls (Bos taurus). Theriogenology 98:41–49. https://doi.org/10.1016/j.theriogenology.2017.04.042
Prakash MA, Kumaresan A, Ebenezer Samuel King JP et al (2021) Comparative transcriptomic analysis of spermatozoa from high- and low-fertile crossbred bulls: implications for fertility prediction. Front Cell Dev Biol 9:1–14. https://doi.org/10.3389/fcell.2021.647717
Pramod RK, Mitra A (2014) In vitro culture and characterization of spermatogonial stem cells on Sertoli cell feeder layer in goat (Capra hircus). J Assist Reprod Genet 31:993–1001. https://doi.org/10.1007/s10815-014-0277-1
Raval NP, Shah TM, George LB, Joshi CG (2019) Insight into bovine (Bos indicus) spermatozoal whole transcriptome profile. Theriogenology 129:8–13. https://doi.org/10.1016/j.theriogenology.2019.01.037
Sahoo B, Guttula PK, Gupta MK (2021) Comparison of spermatozoal RNA extraction methods in goats. Anal Biochem 614:114059. https://doi.org/10.1016/j.ab.2020.114059
Selvaraju S, Parthipan S, Somashekar L et al (2017) Occurrence and functional significance of the transcriptome in bovine (Bos taurus) spermatozoa. Sci Rep 7:1–14. https://doi.org/10.1038/srep42392
Selvaraju S, Parthipan S, Somashekar L et al (2018) Current status of sperm functional genomics and its diagnostic potential of fertility in bovine (Bos taurus). Syst Biol Reprod Med 64:484–501. https://doi.org/10.1080/19396368.2018.1444816
Song Y, Milon B, Ott S et al (2018) A comparative analysis of library prep approaches for sequencing low input translatome samples. BMC Genomics 19:1–16. https://doi.org/10.1186/s12864-018-5066-2
Sun P, Wang Y, Gao T et al (2021) Hsp90 modulates human sperm capacitation via the Erk1/2 and p38 MAPK signaling pathways. Reprod Biol Endocrinol 19:1–11. https://doi.org/10.1186/s12958-021-00723-2
Tan K, Wang X, Zhang Z et al (2016) Downregulation of miR-199a-5p disrupts the developmental potential of in vitro-fertilized mouse blastocysts. Biol Reprod 95:1–9. https://doi.org/10.1095/biolreprod.116.141051
Ureña I, González C, Ramón M et al (2022) Exploring the ovine sperm transcriptome by RNAseq techniques: I effect of seasonal conditions on transcripts abundance. PLoS ONE 17:e0264978. https://doi.org/10.1371/journal.pone.0264978
Wang C, Swerdloff RS (2014) Limitations of semen analysis as a test of male fertility and anticipated needs from newer tests. Fertil Steril 102:1502–1507. https://doi.org/10.1016/j.fertnstert.2014.10.021
Wang X, Liu Q, Xu S et al (2018) Transcriptome dynamics during turbot spermatogenesis predicting the potential key genes regulating male germ cell proliferation and maturation. Sci Rep. https://doi.org/10.1038/s41598-018-34149-5
Wang X, Yang C, Guo F et al (2019) Integrated analysis of mRNAs and long noncoding RNAs in the semen from Holstein bulls with high and low sperm motility. Sci Rep 9:1–10. https://doi.org/10.1038/s41598-018-38462-x
Wen J, Luo Q, Wu Y et al (2020) Integrated analysis of long non-coding RNA and mRNA expression profile in myelodysplastic syndromes. Clin Lab 66:825–834. https://doi.org/10.7754/CLIN.LAB.2019.190939
Xu X, Tan Y, Mao H et al (2020) Analysis of long noncoding RNA and mRNA expression profiles of testes with high and low sperm motility in domestic pigeons (Columba livia). Genes (basel). https://doi.org/10.3390/genes11040349
Zhao W, Ahmed S, Ahmed S et al (2021) Analysis of long non-coding RNAs in epididymis of cattleyak associated with male infertility. Theriogenology 160:61–71. https://doi.org/10.1016/j.theriogenology.2020.10.033
Zhu Z, Li R, Ma G et al (2018) 5’-AMP-activated protein kinase regulates goat sperm functions via energy metabolism in vitro. Cell Physiol Biochem 47:2420–2431. https://doi.org/10.1159/000491616
Acknowledgements
This research project was performed at the Department of Biotechnology and Medical Engineering, National Institute of Technology, Rourkela.
Funding
This work was supported by a grant (#BT/PR40124/BTIS/137/14/2021) from the Department of Biotechnology, Government of India.
Author information
Authors and Affiliations
Contributions
BS performed the experiments and analysed the data. MKG provided the resources and finalized the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sahoo, B., Gupta, M.K. Transcriptome Analysis Reveals Spermatogenesis-Related CircRNAs and LncRNAs in Goat Spermatozoa. Biochem Genet (2023). https://doi.org/10.1007/s10528-023-10520-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10528-023-10520-8