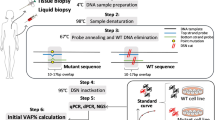
Abstract
PIK3CA mutations have important therapeutic and prognostic implications in various cancer types. However, highly sensitive detection of PIK3CA hotspot mutations in heterogeneous tumor samples remains a challenge in clinical settings. To establish a rapid PCR assay for highly sensitive detection of multiple PIK3CA hotspot mutations. We described a novel melting curve analysis-based assay using looping-out probes that can enrich target mutations in the background of excess wild-type and concurrently reveal the presence of mutations. The analytical and clinical performance of the assay were evaluated. The developed assay could detect 10 PIK3CA hotspot mutations at a mutant allele fraction of 0.05–0.5% within 2 h in a single step. Analysis of 82 breast cancer tissue samples revealed 43 samples with PIK3CA mutations, 28 of which were confirmed by Sanger sequencing. Further testing of 175 colorectal cancer tissue samples showed that 24 samples contained PIK3CA mutations and 19 samples were confirmed by Sanger sequencing. Droplet digital PCR supported that all mutation-containing samples undetected by sequencing contained mutations with a low allele fraction. The rapidity, ease of use, high sensitivity and accuracy make the new assay a potential screening tool for PIK3CA mutations in clinical laboratories.




Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its additional files.
Abbreviations
- PI3Ks:
-
Phosphatidylinositol-3-kinases
- BC:
-
Breast cancer
- CRC:
-
Colorectal cancer
- MAF:
-
Mutant allele fraction
- ARMS:
-
Amplification refractory mutation system
- COLD-PCR:
-
Co-amplification at lower denaturation temperature-polymerase chain reaction
- LNA:
-
Locked nucleic acid
- ddPCR:
-
Droplet digital PCR
- NGS:
-
Next-generation sequencing
- LPO:
-
Looping-out
- gDNA:
-
Genomic DNA
- FFPE:
-
Formalin-fixed paraffin-embedded
- dF/dT:
-
The negative derivative of fluorescence over temperature
- T m :
-
The melting point
- ΔT m :
-
The Tm difference
References
Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, Iwata H, Conte P, Mayer IA, Kaufman B, Yamashita T, Lu YS, Inoue K, Takahashi M, Papai Z, Longin AS, Mills D, Wilke C, Hirawat S, Juric D, Group S-S (2019) Alpelisib for Pik3ca-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 380(20):1929–1940
Ang D, O’Gara R, Schilling A, Beadling C, Warrick A, Troxell ML, Corless CL (2013) Novel method for Pik3ca mutation analysis locked nucleic Acid-Pcr sequencing. J Clin Microbiol 15(3):312–318
Bernard PS, Wittwer CT (2002) Real-time Pcr technology for cancer diagnostics. Clin Chem 48(8):1178–1185
Bidshahri R, Attali D, Fakhfakh K, McNeil K, Karsan A, Won JR, Wolber R, Bryan J, Hughesman C, Haynes C (2016) Quantitative detection and resolution of Braf V600 status in colorectal cancer using droplet digital Pcr and a novel wild-type negative assay. J Mol Diagn 18(2):190–204
Chen L, Yang L, Yao L, Kuang XY, Zuo WJ, Li S, Qiao F, Liu YR, Cao ZG, Zhou SL, Zhou XY, Yang WT, Shi JX, Huang W, Hu X, Shao ZM (2018) Characterization of Pik3ca and Pik3r1 somatic mutations in chinese breast cancer patients. Nat Commun 9(1):1357
Chen Z, Wang C, Dong H, Wang X, Gao F, Zhang S, Zhang X (2020) Aspirin has a better effect on Pik3ca mutant colorectal cancer cells by Pi3k/Akt/raptor pathway. Mol Med 26(1):14
Cizkova M, Dujaric ME, Lehmann-Che J, Scott V, Tembo O, Asselain B, Pierga JY, Marty M, de Cremoux P, Spyratos F, Bieche I (2013) Outcome impact of Pik3ca mutations in Her2-positive breast cancer patients treated with trastuzumab. Br J Cancer 108(9):1807–1809
Decraene C, Silveira AB, Bidard FC, Vallee A, Michel M, Melaabi S, Vincent-Salomon A, Saliou A, Houy A, Milder M, Lantz O, Ychou M, Denis MG, Pierga JY, Stern MH, Proudhon C (2018) Multiple hotspot mutations scanning by single droplet digital Pcr. Clin Chem 64(2):317–328
Deng L, Zhu X, Sun Y, Wang J, Zhong X, Li J, Hu M, Zheng H (2019) Prevalence and prognostic role of Pik3ca/Akt1 mutations in chinese breast cancer patients. Cancer Res Treat 51(1):128–140
Do H, Dobrovic A (2015) Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem 61(1):64–71
Fusco N, Malapelle U, Fassan M, Marchio C, Buglioni S, Zupo S, Criscitiello C, Vigneri P, Dei Tos AP, Maiorano E, Viale G (2021) Pik3ca mutations as a molecular target for hormone receptor-positive, her2-negative metastatic breast cancer. Front Oncol 11:644737
Ghalamkari S, Khosravian F, Mianesaz H, Kazemi M, Behjati M, Hakimian SM, Salehi M (2019) A comparison between full-cold Pcr/Hrm and Pcr sequencing for detection of mutations in exon 9 of Pik3ca in breast cancer patients. Appl Biochem Biotechnol 187(3):975–983
Gradishar WJ, Moran MS, Abraham J, Aft R, Agnese D, Allison KH, Anderson B, Burstein HJ, Chew H, Dang C, Elias AD, Giordano SH, Goetz MP, Goldstein LJ, Hurvitz SA, Isakoff SJ, Jankowitz RC, Javid SH, Krishnamurthy J, Leitch M, Lyons J, Mortimer J, Patel SA, Pierce LJ, Rosenberger LH, Rugo HS, Sitapati A, Smith KL, Smith ML, Soliman H, Stringer-Reasor EM, Telli ML, Ward JH, Wisinski KB, Young JS, Burns J, Kumar R (2022) Breast cancer, version 3.2022, Nccn clinical practice guidelines in oncology. J Natl Compr Canc Netw 20(6):691–722
Guo F, Gong H, Zhao H, Chen J, Zhang Y, Zhang L, Shi X, Zhang A, Jin H, Zhang J, He Y (2018) Mutation status and prognostic values of Kras, Nras, Braf and Pik3ca in 353 Chinese colorectal cancer patients. Sci Rep 8(1):6076
Harle A, Lion M, Lozano N, Husson M, Harter V, Genin P, Merlin JL (2013) Analysis of Pik3ca Exon 9 and 20 mutations in breast cancers using Pcr-Hrm and Pcr-Arms: correlation with clinicopathological criteria. Oncol Rep 29(3):1043–1052
He Y, Van’t Veer LJ, Mikolajewska-Hanclich I, van Velthuysen M-LF, Zeestraten ECM, Nagtegaal ID, van de Velde CJH, Marijnen CAM (2009) Pik3ca mutations predict local recurrences in rectal cancer patients. Clin Cancer Res 15(22):6956–6962
Huang Q, Liu Z, Liao Y, Chen X, Zhang Y, Li Q (2011) Multiplex fluorescence melting curve analysis for mutation detection with dual-labeled. Self-Quenched Probes Plos One 6(4):e19206
Hurst CD, Zuiverloon TC, Hafner C, Zwarthoff EC, Knowles MA (2009) A snapshot assay for the rapid and simple detection of four common hotspot codon mutations in the Pik3ca gene. BMC Res Notes 2:66
Kim ST, Lira M, Deng SB, Lee S, Park YS, Lim HY, Kang WK, Mao M, Heo JS, Kwon W, Jang KT, Lee J, Park JO (2015) Pik3ca mutation detection in metastatic biliary cancer using cell-free DNA. Oncotarget 6(37):40026–40035
Kwon MJ, Lee SE, Kang SY, Choi YL (2011) Frequency of Kras, Braf, and Pik3ca mutations in advanced colorectal cancers: comparison of peptide nucleic acid-mediated Pcr clamping and direct sequencing in formalin-fixed. Paraffin-Embedded Tissue Pathol Res Pract 207(12):762–768
Liang B, Tan YJ, Li Z, Tian XS, Du C, Li H, Li GL, Yao XY, Wang ZA, Xu Y, Li QG (2018) Highly sensitive detection of isoniazid heteroresistance in mycobacterium tuberculosis by deepmelt assay. J Clin Microbiol 56(2):e01239-e1317
Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt JA, Fuchs CS, Ogino S (2012) Prognostic role of Pik3ca mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res 18(8):2257–2268
Ligresti G, Militello L, Steelman LS, Cavallaro A, Basile F, Nicoletti F, Stivala F, McCubrey JA, Libra M (2009) Pik3ca mutations in human solid tumors: role in sensitivity to various therapeutic approaches. Cell Cycle 8(9):1352–1358
Loibl S, von Minckwitz G, Schneeweiss A, Paepke S, Lehmann A, Rezai M, Zahm DM, Sinn P, Khandan F, Eidtmann H, Dohnal K, Heinrichs C, Huober J, Pfitzner B, Fasching PA, Andre F, Lindner JL, Sotiriou C, Dykgers A, Guo S, Gade S, Nekljudova V, Loi S, Untch M, Denkert C (2014) Pik3ca mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (Her2) therapy in primary Her2-overexpressing breast cancer. J Clin Oncol 32(29):3212–3220
Luo JD, Chan EC, Shih CL, Chen TL, Liang Y, Hwang TL, Chiou CC (2006) Detection of rare mutant K-Ras DNA in a single-tube reaction using peptide nucleic acid as both Pcr clamp and sensor probe. Nucleic Acids Res 34(2):e12
Markou A, Farkona S, Schiza C, Efstathiou T, Kounelis S, Malamos N, Georgoulias V, Lianidou E (2014) Pik3ca mutational status in circulating tumor cells can change during disease recurrence or progression in patients with breast cancer. Clin Cancer Res 20(22):5823–5834
Martinez-Saez O, Chic N, Pascual T, Adamo B, Vidal M, Gonzalez-Farre B, Sanfeliu E, Schettini F, Conte B, Braso-Maristany F, Rodriguez A, Martinez D, Galvan P, Rodriguez AB, Martinez A, Munoz M, Prat A (2020) Frequency and spectrum of Pik3ca somatic mutations in breast cancer. Breast Cancer Res 22(1):45
Nakauchi C, Kagara N, Shimazu K, Shimomura A, Naoi Y, Shimoda M, Kim SJ, Noguchi S (2016) Detection of Tp53/Pik3ca mutations in cell-free plasma DNA from metastatic breast cancer patients using next generation sequencing. Clin Breast Cancer 16(5):418–423
Ney JT, Froehner S, Roesler A, Buettner R, Merkelbach-Bruse S (2012) High-resolution melting analysis as a sensitive prescreening diagnostic tool to detect Kras, Braf, Pik3ca, and Akt1 mutations in formalin-fixed. Paraffin-Embedded Tissues Arch Pathol Lab Med 136(9):983–992
Ogino S, Nosho K, Kirkner GJ, Shima K, Irahara N, Kure S, Chan AT, Engelman JA, Kraft P, Cantley LC, Giovannucci EL, Fuchs CS (2009) Pik3ca mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol 27(9):1477–1484
Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, Anagnostou V, Fiksel J, Cristiano S, Papp E, Speir S, Reinert T, Orntoft MBW, Woodward BD, Murphy D, Parpart-Li S, Riley D, Nesselbush M, Sengamalay N, Georgiadis A, Li QK, Madsen MR, Mortensen FV, Huiskens J, Punt C, van Grieken N, Fijneman R, Meijer G, Husain H, Scharpf RB, Diaz LA, Jones S, Angiuoli S, Orntoft T, Nielsen HJ, Andersen CL, Velculescu VE (2017) Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aan2415
Piscuoglio S, Burke KA, Ng CK, Papanastasiou AD, Geyer FC, Macedo GS, Martelotto LG, de Bruijn I, De Filippo MR, Schultheis AM, Ioris RA, Levine DA, Soslow RA, Rubin BP, Reis-Filho JS, Weigelt B (2016) Uterine adenosarcomas are mesenchymal neoplasms. J Pathol 238(3):381–388
Pont-Kingdon G, Lyon E (2005) Direct molecular haplotyping by melting curve analysis of hybridization probes: beta 2-adrenergic receptor haplotypes as an example. Nucleic Acids Res 33(10):e89
Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE (2004) High frequency of mutations of the Pik3ca gene in human cancers. Science 304(5670):554
Turner NC, Alarcon E, Armstrong AC, Philco M, Lopez Chuken YA, Sablin MP, Tamura K, Gomez Villanueva A, Perez-Fidalgo JA, Cheung SYA, Corcoran C, Cullberg M, Davies BR, de Bruin EC, Foxley A, Lindemann JPO, Maudsley R, Moschetta M, Outhwaite E, Pass M, Rugman P, Schiavon G, Oliveira M (2019) Beech: a dose-finding run-in followed by a randomised phase ii study assessing the efficacy of Akt inhibitor capivasertib (Azd5363) combined with paclitaxel in patients with estrogen receptor-positive advanced or metastatic breast cancer, and in a Pik3ca mutant sub-population. Ann Oncol 30(5):774–780
Vivanco I, Sawyers CL (2002) The Phosphatidylinositol 3-kinase Akt pathway in human cancer. Nat Rev Cancer 2(7):489–501
Vorkas PA, Poumpouridou N, Agelaki S, Kroupis C, Georgoulias V, Lianidou ES (2010) Pik3ca hotspot mutation scanning by a novel and highly sensitive high-resolution small amplicon melting analysis method. J Mol Diagn 12(5):697–704
Zhang QY, Cheng WX, Li WM, Au W, Lu YY (2014) Occurrence of low frequency Pik3ca and Akt2 mutations in gastric cancer. Mutat Res-Fund Mol M 769:108–112
Zhao L, Vogt PK (2008) Helical domain and kinase domain mutations in P110 alpha of phosphatidylinositol 3-kinase induce gain of function by different mechanisms. P Natl Acad Sci USA 105(7):2652–2657
Funding
This study was supported by National Natural Science Foundation of China (Grant No. 81672110).
Author information
Authors and Affiliations
Contributions
BHX, YPL and DJL design and performed experiments, analyzed data and co-wrote this paper. YSZ, RFN and GQS collected samples and the data. QYH and QGL provided financial support and supervised the study proposal. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical Approval
Approval of the research protocol by an Institutional Reviewer Board: this study was approved by the medical ethics committee of Xiamen University (approval No. XDYX2021017).
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent to Publish
The authors affirm that human research participants provided informed consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, B., Lan, Y., Luo, D. et al. Highly Sensitive Detection of PIK3CA Mutations by Looping-Out Probes-Based Melting Curve Analysis. Biochem Genet 62, 77–94 (2024). https://doi.org/10.1007/s10528-023-10408-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-023-10408-7