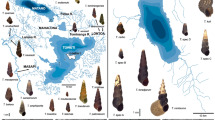
Abstract
To investigate the genetic diversity and genetic variations of four wild (Geoje, Jinhae, Yeosu, and Boryeong) and two hatchery (Goheung and Geoje) populations of purplish Washington clam (Saxidomus purpuratus), 421 bp sequences of the mitochondrial cytochrome c oxidase subunit I (COI) gene were analyzed. A total of 149 haplotypes were identified from 358 individuals from the four wild and two hatchery populations with 109 substitutions. The genetic diversity of the wild populations and Geoje hatchery population were high, whereas the total number of haplotypes, population-specific haplotypes, and haplotype diversity were comparatively low in the Goheung hatchery population. The fixation index (FST) indicated that there was no significant genetic difference between the four wild populations. However, the Goheung hatchery population was significantly different from that of the Geoje hatchery, exhibiting the most pronounced difference, and two wild populations (Jinhae and Yeosu). The low genetic diversity indices exhibited by the Goheung hatchery population might have resulted from farm propagation using a limited parental stock. Therefore, to maintain genetic diversity, a proper breeding management program using more progenitors is required in hatcheries, in addition to regular monitoring of both hatchery and wild populations.




Similar content being viewed by others
Availability of Data and Material
All the haplotypes observed in the present study were deposited to the GenBank with accession numbers MT863090-MT863238.
References
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190
Allendorf FW, Ryman N (1987) Genetic management of hatchery stocks. In: Ryman N, Utter F (eds) Population genetics and fishery management. University of Washington Press, Seattle, pp 141–159
Allendorf FW, Phelps SR (1980) Loss of genetic variation in a hatchery stock of cutthroat trout. Trans Am Fish Soc 109:537–543
Aung O, Nguyen TT, Poompuang S, Kamonrat W (2010) Microsatellite DNA markers revealed genetic population structure among captive stocks and wild populations of mrigal, Cirrhinus cirrhosus in Myanmar. Aquaculture 299:37–43
Barrett SC, Charlesworth D (1991) Effects of a change in the level of inbreeding on the genetic load. Nature 352:522–524
Bekessy SA, Ennos RA, Burgman MA, Newton AC, Ades PK (2003) Neutral DNA markers fail to detect genetic divergence in an ecologically important trait. Biol Conserv 110:267–275
Blin N, Stafford DW (1976) A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res 3:2303–2308
Cho ES, Jung CG, Sohn SG, Kim CW, Han SJ (2007) Population genetic structure of the ark shell scapharca broughtonii schrenck from korea, china, and russia based on COI gene sequences. Mar Biotechnol 9:203–216
Cho ES, Seo YI, Suh YS (2012) Genetic analysis of the purplish Washington clam (Saxidomus purpuratus Sowerby) of Korean coastal waters. J Environ Biol 34:613–621
Choi YS, Kim IS, Bang JD (2001) Studies on the development of techniques on seedling production of Saxidomus purpuratus. Report of West Sea Fisheries Research Institute, National Fisheries Research and Development Institute (In Korean)
Evans B, Bartlett J, Sweijd N, Cook P, Elliott NG (2004) Loss of genetic variation at microsatellite loci in hatchery produced abalone in Australia (Haliotis rubra) and South Africa (Haliotis midae). Aquaculture 233:109–127
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Eyre-Walker A, Awadalla P (2001) Does human mtDNA recombine? J Mol Evol 53:430–435
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–299
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925
Grant WS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered 89:415–426
Gwak WS, Nakayama K (2011) Genetic variation of hatchery and wild stocks of the pearl oyster Pinctada fucata martensii (Dunker, 1872), assessed by mitochondrial DNA analysis. Aquacult Int 19:585–591
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Harpending RC (1994) Signature of ancient population growth in a low resolution mitochondrial DNA mismatch distribution. Hum Biol 66:591–600
Hewitt GM (2000) The genetic legacy of the Quaternary ice ages. Nature 405:907–913
Hsu TH, Gwo JC (2017) Genetic diversity and stock identification of small abalone (Haliotis diversicolor) in Taiwan and Japan. PLoS ONE 12:e0179818
Irvin SD, Wetterstrand KA, Hutter CM, Aquadro CF (1998) Genetic variability and differentiation at microsatellite loci in Droscphila simulans: evidence for founder effects in New World populations. Genetics 150:777–790
Ji CH, Gu JJ, Mao RX, Zhu XP, Sun XW (2009) Analysis of genetic diversity among wild silver carp (Hypophthamlichthys molitrix) populations in the Yangtze. Heilongjiang and Pearl Rivers using microsatellite markers. J Fish China 33:354–371
Jin YG, Oh BS, Jung CK, Kim TI, Park MW (2011) Survival and growth of the purplish Washington clam, Saxidomus purpuratus spat sowed in bottom and intermediate culture. Korean J Malacol 27:199–204
Kim SK, Park KY, Jang GN, Choi YS, Kim IS (2000) Studies on the development of techniques on seedling production of Saxidomus purpuratus. Report of West Sea Fisheries Research Institute, National Fisheries Research and Development Institute, Korea, pp 343–352. (in Korean)
Kim SK, Park KY, Jang GN, Kim DJ, Seo HC (2001) Studies on the ecological aspects and gametogenesis of Saxidomus purpuratus (Sowerby) in the Yellow Sea area. Bull Natl Fish Res Dev Inst Korea 59:152–158 (in Korean)
Kim YH, Kwon DH, Chang DS, Kim JB, Kim ST, Ryu DK (2007) Stock assessment of purplish Washington clam, Saxidomus purpuratus in the southern coastal waters of Korea. Korean J Malacol 23:31–38
Korean Fisheries Yearbook (2017) www.fips.go.kr
KOSIS (2016) Fishery, species and fishing method statistics [Internet]. c2006/2015. Korea: Korean statistical information service. . Available at: kosis.kr. Accessed 11 Mar 2016
Leber KM, Brennan NP, Arce SM (1995) Marine enhancement with striped mullet: are hatchery releases replenishing or displacing wild stocks. In: American Fisheries Society Symposium, vol 15, pp 376–387
Lee SK, Chang DS, Kim JB, Park MA (2013) Reproduction study of purplish Washington clam, Saxidomus purpuratus in Jinhae bay, Korea: Spawning and shell length at 50% sexual maturity. J Korean Soc Fish Technol 49:449–458
Li Q, Park C, Endo T, Kijima A (2004) Loss of genetic variation at microsatellite loci in hatchery strains of the Pacific abalone (Haliotis discus hannai). Aquaculture 235:207–222
Liu JX, Gao TX, Wu SF, Zhang YP (2007) Pleistocene isolation in the Northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus (Temminck & Schlegel, 1845). Mol Ecol 16:275–288
Ovenden JR, Leigh GM, Blower DC, Jones AT, Moore A, Bustamante C, Buckworth RC, Bennet MB, Dudgeon CL (2016) Can estimates of genetic effective population size contribute to fisheries stock assessments? J Fish Biol 89:2505–2518
Park KI, Choi JW, Choi KS (2005) Development of a Saxidomus purpuratus (mollusca: bivalvia) egg-specific antibody for the quantification of eggs using an enzyme-linked immunosorbent assay. J Shellfish Res 24:1079–1085
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Seed R (1969) The ecology of Mytilus edulis L. (Lamelibrabchiata) on exposed rocky shores. 1. Breed Settl Oecol 3:277–316
Shen YB, Li JL, Feng BB (2009) Genetic analysis of cultured and wild populations of Mytilus coruscus based on mitochondrial DNA. Zool Res 30:240–246
Spielman D, Brook BW, Briscoe DA, Frankham R (2004) Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet 5:439–448
Stopar K, Ramsak A, Trontelj P, Malej A (2010) Lack of genetic structure in the jellyfish Pelagia noctiluca (Cnidaria: Scyphozoa: Semaeostomeae) across European seas. Mol Phylogenet Evol 57:417–428
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Teacher AGF, Griffiths DJ (2011) Hapstar: automated haplotype network layout and visualization. Mol Ecol Resour 11:151–153
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Wang J, Tsang LM, Dong Y (2015) Causations of phylogeographic barrier of some rocky shore species along the Chinese coastline. BMC Evol Biol 15:114
Wei LP, Shu YF, Guan F, Han YP, Yu ZR (1982) A preliminary survey on the biology of Saxidomus purpuratus. J Fish China 6:1–8 (in Chinese with English abstract)
Xiao J (2009) Assessing genetic variation within and among native populations and hatchery stocks of Crassostrea ariakensis using microsatellite markers [Ph.D. 3353206]. The College of William and Mary
Zheng JH, Nie HT, Yang F, Yan XW (2019) Genetic variation and population structure of different geographical populations of Meretrix petechialis based on mitochondrial gene COI. J Genet 98(3):1–9
Acknowledgements
We thank Jongyul Park for assistance of experiment. This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2012R1A1A2004830).
Funding
This work was supported by the National Research Foundation of Korea Grant funded by the Korean Government (NRF-2012R1A1A2004830).
Author information
Authors and Affiliations
Contributions
W-SG contributed to conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; roles/writing—original draft; and writing—review and editing. AR participated in the writing of the original draft of the manuscript; software; and data curation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gwak, WS., Roy, A. Genetic Diversity and Variation in Mitochondrial COI Gene in Wild and Hatchery Populations of Saxidomus purpuratus. Biochem Genet 60, 969–986 (2022). https://doi.org/10.1007/s10528-021-10137-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-021-10137-9