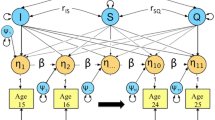
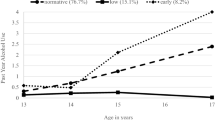
Abstract
Contemporary genome-wide association study (GWAS) methods typically do not account for variability in genetic effects throughout development. We applied genomic structural equation modeling to combine developmentally-informative phenotype data and GWAS to create polygenic scores (PGS) for alcohol use frequency that are specific to developmental stage. Longitudinal cohort studies targeted for gene-identification analyses include the Collaborative Study on the Genetics of Alcoholism (adolescence n = 1,118, early adulthood n = 2,762, adulthood n = 5,255), the National Longitudinal Study of Adolescent to Adult Health (adolescence n = 3,089, early adulthood n = 3,993, adulthood n = 5,149), and the Avon Longitudinal Study of Parents and Children (ALSPAC; adolescence n = 5,382, early adulthood n = 3,613). PGS validation analyses were conducted in the COGA sample using an alternate version of the discovery analysis with COGA removed. Results suggest that genetic liability for alcohol use frequency in adolescence may be distinct from genetic liability for alcohol use frequency later in developmental periods. The age-specific PGS predicts an increase of 4 drinking days per year per PGS standard deviation when modeled separately from the common factor PGS in adulthood. The current work was underpowered at all steps of the analysis plan. Though small sample sizes and low statistical power limit the substantive conclusions that can be drawn regarding these research questions, this work provides a foundation for future genetic studies of developmental variability in the genetic underpinnings of alcohol use behaviors and genetically-informed, age-matched phenotype prediction.






Similar content being viewed by others
Data Availability
All data sources are described in the manuscript. No new data were collected. Only data from existing studies or study cohorts were analyzed. Add Health genetic data obtained through dbGaP (Study Accession: phs001367.v1.p1). Instructions on gaining access to Add Health restricted use data can be found at: https://data.cpc.unc.edu/projects/2/view. COGA genetic data available through dbGaP (Study Accession: phs000763.v1.p1). Instructions for access to ALSPAC data available at: http://www.bristol.ac.uk/alspac/researchers/access/.
Code Availability
No custom software was developed in this study. All code is available by request from the corresponding author.
References
Aliev F, Wetherill L, Bierut L, Bucholz KK, Edenberg H, Foroud T, Dick DM (2015) Genes Associated with Alcohol Outcomes Show Enrichment of effects with Broad Externalizing and Impulsivity Phenotypes in an Independent sample. J Stud Alcohol Drugs 76:38–46
Altshuler DM, Gibbs RA, Peltonen L et al (2010) Integrating common and rare genetic variation in diverse human populations. Nature 467:52–58. https://doi.org/10.1038/nature09298
Barr PB, Mallard TT, Sanchez-Roige S, Poore HE, Linnér RK, Waldman ID, Palmer AA, Harden KP, Agrawal A, Dick DM (2022) Parsing genetically influenced risk pathways: genetic loci impact problematic alcohol use via externalizing and specific risk. Transl Psychiatry 12:1–8. https://doi.org/10.1038/s41398-022-02171-x
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting Linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Begleiter H (1995) The collaborative study on the Genetics of Alcoholism. Alcohol Health Res World 19:228–236
Benjamini Y, Hochberg Y (1995) Controlling the false Discovery rate: a practical and powerful Approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 57:289–300. https://doi.org/10.1111/j.2517-6161.1995.tb02031.x
Boyd A, Golding J, Macleod J, Lawlor DA, Fraser A, Henderson J, Molloy L, Ness A, Ring S, Davey Smith G (2013) Cohort Profile: the ‘Children of the 90s’—the index offspring of the Avon Longitudinal Study of parents and children. Int J Epidemiol 42:111–127. https://doi.org/10.1093/ije/dys064
Bryazka D, Reitsma MB, Griswold MG et al (2022) Population-level risks of alcohol consumption by amount, geography, age, sex, and year: a systematic analysis for the global burden of Disease Study 2020. The Lancet 400:185–235. https://doi.org/10.1016/S0140-6736(22)00847-9
Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Reich T, Schmidt I, Schuckit MA (1994) A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol 55:149–158. https://doi.org/10.15288/jsa.1994.55.149
Bucholz KK, McCutcheon VV, Agrawal A, Dick DM, Hesselbrock VM, Kramer JR, Kuperman S, Nurnberger JI, Salvatore JE, Schuckit MA, Bierut LJ, Foroud TM, Chan G, Hesselbrock M, Meyers JL, Edenberg HJ, Porjesz B (2017) Comparison of parent, peer, psychiatric, and cannabis use influences across stages of offspring alcohol involvement: evidence from the COGA prospective study. Alcohol Clin Exp Res 41:359–368. https://doi.org/10.1111/acer.13293
Bulik-Sullivan BK, Finucane HK, Anttila V, Gusev A, Day FR, Loh P-R, ReproGen, Consortium, Psychiatric Genomics Consortium, Genetic Consortium for Anorexia Nervosa of the Wellcome Trust Case Control Consortium, Duncan L, Perry JRB, Patterson N, Robinson EB, Daly MJ, Price AL, Neale BM (2015) An atlas of genetic correlations across human Diseases and traits. Nat Genet 47:1236–1241. https://doi.org/10.1038/ng.3406
Bulik-Sullivan BK, Loh P-R, Finucane HK, Ripke S, Yang J, Patterson N, Daly MJ, Price AL, Neale BM (2015) LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet 47:291–295. https://doi.org/10.1038/ng.3211
Chen Z, Boehnke M, Wen X, Mukherjee B (2021) Revisiting the genome-wide significance threshold for common variant GWAS. G3 Genes|Genomes|Genetics. 11:jkaa056. https://doi.org/10.1093/g3journal/jkaa056
Chong AHW, Mitchell RE, Hemani G, Davey Smith G, Yolken RH, Richmond RC, Paternoster L (2021) Genetic Analyses of Common Infections in the Avon Longitudinal Study of parents and children Cohort. Front Immunol 12
Dawson DA (2000) US low-risk drinking guidelines: an examination of four alternatives. Alcoholism: Clin Experimental Res 24:1820–1829. https://doi.org/10.1111/j.1530-0277.2000.tb01986.x
Dick DM, Bierut L, Hinrichs A, Fox L, Bucholz KK, Kramer J, Kuperman S, Hesselbrock V, Schuckit M, Almasy L, Tischfield J, Porjesz B, Begleiter H, Nurnberger J, Xuei X, Edenberg HJ, Foroud T (2006) The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav Genet 36:577–590. https://doi.org/10.1007/s10519-005-9041-8
Dick DM, Latendresse SJ, Lansford JE, Budde JP, Goate A, Dodge KA, Pettit GS, Bates JE (2009) Role of GABRA2 in trajectories of externalizing Behavior Across Development and evidence of moderation by parental monitoring. Arch Gen Psychiatry 66:649–657. https://doi.org/10.1001/archgenpsychiatry.2009.48
Dick DM, Balcke E, McCutcheon V, Francis M, Kuo S, Salvatore J, Meyers J, Schuckit M, Hesselbrock V, Edenberg HJ, Kuperman S, Kramer J, Bucholz K COGA Collaborators (in press) the collaborative study on the Genetics of Alcoholism: 2. Data Collection, Sample characteristics, and key findings. G2B
Edwards AC, Kendler KS (2013) Alcohol consumption in men is influenced by qualitatively different genetic factors in adolescence and adulthood. Psychol Med 43:1857–1868. https://doi.org/10.1017/S0033291712002917
Elam KK, Ha T, Neale Z, Aliev F, Dick D, Lemery-Chalfant K (2021) Age varying polygenic effects on alcohol use in African americans and European americans from adolescence to adulthood. Sci Rep 11:22425. https://doi.org/10.1038/s41598-021-01923-x
Fowler T, Lifford K, Shelton K, Rice F, Thapar A, Neale MC, McBride A, van den Bree MBM (2007) Exploring the relationship between genetic and environmental influences on initiation and progression of substance use. Addiction 102:413–422. https://doi.org/10.1111/j.1360-0443.2006.01694.x
Fraser A, Macdonald-Wallis C, Tilling K, Boyd A, Golding J, Davey Smith G, Henderson J, Macleod J, Molloy L, Ness A, Ring S, Nelson SM, Lawlor DA (2013) Cohort Profile: the Avon Longitudinal Study of parents and children: ALSPAC mothers cohort. Int J Epidemiol 42:97–110. https://doi.org/10.1093/ije/dys066
Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW (2019) Polygenic prediction via bayesian regression and continuous shrinkage priors. Nat Commun 10:1–10. https://doi.org/10.1038/s41467-019-09718-5
Genz A, Bretz F (2009) Computation of Multivariate Normal and t probabilities. Springer Berlin, Heidelberg
Genz A, Bretz F, Miwa T, Mi X, Leisch F, Scheipl F, Hothorn T (2021) mvtnorm: Multivariate Normal and t Distributions. Version R package version 1.1-3URL https://CRAN.R-project.org/package=mvtnorm
Gillespie NA, Gentry AE, Kirkpatrick RM, Reynolds CA, Mathur R, Kendler KS, Maes HH, Webb BT, Peterson RE (2022) Determining the stability of genome-wide factors in BMI between ages 40 to 69 years. PLoS Genet 18:e1010303. https://doi.org/10.1371/journal.pgen.1010303
Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD, Ip HF, Marioni RE, McIntosh AM, Deary IJ, Koellinger PD, Harden KP, Nivard MG, Tucker-Drob EM (2019) Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nat Hum Behav 3:513–525. https://doi.org/10.1038/s41562-019-0566-x
Harris KM (2013) The Add Health Study: Design and Accomplishments
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG (2009) Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377–381. https://doi.org/10.1016/j.jbi.2008.08.010
Heath AC, Meyer J, Eaves LJ, Martin NG (1991) The inheritance of alcohol consumption patterns in a general population twin sample: I. Multidimensional scaling of quantity/frequency data. J Stud Alcohol 52:345–352. https://doi.org/10.15288/jsa.1991.52.345
Highland HM, Avery CL, Duan Q, Li Y, Harris KM (2018) Quality Control Analysis of Add Health GWAS Data. Carolina Population Center, Chapel Hill, NC
Howard DM, Adams MJ, Clarke T-K, Hafferty JD, Gibson J, Shirali M, Coleman JRI, Hagenaars SP, Ward J, Wigmore EM, Alloza C, Shen X, Barbu MC, Xu EY, Whalley HC, Marioni RE, Porteous DJ, Davies G, Deary IJ, Hemani G, Berger K, Teismann H, Rawal R, Arolt V, Baune BT, Dannlowski U, Domschke K, Tian C, Hinds DA, 23andMe Research Team, Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium, Trzaskowski M, Byrne EM, Ripke S, Smith DJ, Sullivan PF, Wray NR, Breen G, Lewis CM, McIntosh AM (2019) Genome-wide meta-analysis of depression identifies 102 Independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci 22:343–352. https://doi.org/10.1038/s41593-018-0326-7
Huibregtse BM, Corley RP, Wadsworth SJ, Vandever JM, DeFries JC, Stallings MC (2016) A longitudinal adoption study of Substance Use Behavior in Adolescence. Twin Res Hum Genet 19:330–340. https://doi.org/10.1017/thg.2016.35
Johnson EC, Salvatore JE, Lai D, Merikangas AK, Nurnberger JI, Tischfield JA, Xuei X, Kamarajan C, Wetherill L, Collaborators COGA, Rice J, Kramer JR, Kuperman S, Foroud T, Slesinger PA, Goate AM, Porjesz B, Dick DM, Edenberg HJ, Agrawal A (in press) The Collaborative Study on the Genetics of Alcoholism: 4. Genetics. G2B
Kandaswamy R, Allegrini A, Plomin R, von Stumm S (2021) Predictive validity of genome-wide polygenic scores for alcohol use from adolescence to young adulthood. Drug Alcohol Depend 219:108480. https://doi.org/10.1016/j.drugalcdep.2020.108480
Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, Lebreton M, Tino SP, Abdellaoui A, Hammerschlag AR, Nivard MG, Okbay A, Rietveld CA, Timshel PN, Trzaskowski M, de Vlaming R, Zünd CL, Bao Y, Buzdugan L, Caplin AH, Chen C-Y, Eibich P, Fontanillas P, Gonzalez JR, Joshi PK, Karhunen V, Kleinman A, Levin RZ, Lill CM, Meddens GA, Muntané G, Sanchez-Roige S, van Rooij FJ, Taskesen E, Wu Y, Zhang F, Auton A, Boardman JD, Clark DW, Conlin A, Dolan CC, Fischbacher U, Groenen PJF, Harris KM, Hasler G, Hofman A, Ikram MA, Jain S, Karlsson R, Kessler RC, Kooyman M, MacKillop J, Männikkö M, Morcillo-Suarez C, McQueen MB, Schmidt KM, Smart MC, Sutter M, Thurik AR, Uitterlinden AG, White J, de Wit H, Yang J, Bertram L, Jan Bonder M, Boomsma DI, Esko T, Fehr E, Hinds DA, Johannesson M, Kumari M, Laibson D, Magnusson PKE, Meyer MN, Navarro A, Palmer AA, Pers TH, Posthuma D, Schunk D, Stein MB, Svento R, Tiemeier H, Timmers PRHJ, Turley P, Ursano RJ, Wagner GG, Wilson JF, Gratten J, Kutalik Z, Lee JJ, Cesarini D, Benjamin DJ, Koellinger PD, Beauchamp JP (2019) Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet 51:245–257. https://doi.org/10.1038/s41588-018-0309-3
Karlsson Linnér R, Mallard TT, Barr PB, Sanchez-Roige S, Madole JW, Driver MN, Poore HE, Grotzinger AD, Tielbeek JJ, Johnson EC, Liu M, Zhou H, Kember RL, Pasman JA, Verweij KJH, Liu DJ, Vrieze S, Collaborators C, Kranzler HR, Gelernter J, Harris KM, Tucker-Drob EM, Waldman I, Palmer AA, Harden KP, Koellinger PD, Dick DM (2020) Multivariate genomic analysis of 1.5 million people identifies genes related to addiction, antisocial behavior, and health. bioRxiv 2020.10.16.342501. https://doi.org/10.1101/2020.10.16.342501
Kendler KS, Schmitt E, Aggen SH, Prescott CA (2008) Genetic and environmental influences on alcohol, caffeine, cannabis, and nicotine use from early adolescence to middle adulthood. Arch Gen Psychiatry 65:674–682. https://doi.org/10.1001/archpsyc.65.6.674
Kendler KS, Gardner C, Dick DM (2011) Predicting alcohol consumption in adolescence from alcohol-specific and general externalizing genetic risk factors, key environmental exposures and their interaction. Psychol Med 41:1507–1516. https://doi.org/10.1017/S003329171000190X
Kranzler HR, Zhou H, Kember RL, Smith RV, Justice AC, Damrauer S, Tsao PS, Klarin D, Baras A, Reid J, Overton J, Rader DJ, Cheng Z, Tate JP, Becker WC, Concato J, Xu K, Polimanti R, Zhao H, Gelernter J (2019) Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun 10:1–11. https://doi.org/10.1038/s41467-019-09480-8
Kranzler HR, Zhou H, Kember RL (2022) Identifying and reducing Bias in genome-wide Association studies of Alcohol-related traits. AJP 179:14–16. https://doi.org/10.1176/appi.ajp.2021.21111107
Kuznetsova A, Brockhoff PB, Christensen RHB (2017) lmerTest Package: tests in Linear mixed effects models. J Stat Softw 82:1–26. https://doi.org/10.18637/jss.v082.i13
Lai D, Wetherill L, Bertelsen S, Carey CE, Kamarajan C, Kapoor M, Meyers JL, Anokhin AP, Bennett DA, Bucholz KK, Chang KK, De Jager PL, Dick DM, Hesselbrock V, Kramer J, Kuperman S, Nurnberger JI, Raj T, Schuckit M, Scott DM, Taylor RE, Tischfield J, Hariri AR, Edenberg HJ, Agrawal A, Bogdan R, Porjesz B, Goate AM, Foroud T (2019) Genome-wide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes Brain Behav 18:e12579. https://doi.org/10.1111/gbb.12579
Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet 11:241–247. https://doi.org/10.1038/ng1195-241
Liu M, Jiang Y, Wedow R et al (2019) Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of Tobacco and alcohol use. Nat Genet 51:237–244. https://doi.org/10.1038/s41588-018-0307-5
Long EC, Verhulst B, Aggen SH, Kendler KS, Gillespie NA (2017) Contributions of genes and environment to Developmental Change in Alcohol Use. Behav Genet 47:498–506. https://doi.org/10.1007/s10519-017-9858-y
Lüdecke D, Ben-Shachar MS, Patil I, Waggoner P, Makowski D (2021) Performance: an R Package for Assessment, comparison and testing of statistical models. J Open Source Softw 6:3139. https://doi.org/10.21105/joss.03139
Mallard TT, Karlsson Linnér R, Grotzinger AD, Sanchez-Roige S, Seidlitz J, Okbay A, de Vlaming R, Meddens SFW, Palmer AA, Davis LK, Tucker-Drob EM, Kendler KS, Keller MC, Koellinger PD, Harden KP (2022a) Multivariate GWAS of psychiatric disorders and their cardinal symptoms reveal two dimensions of cross-cutting genetic liabilities. Cell Genomics 2:100140. https://doi.org/10.1016/j.xgen.2022.100140
Mallard TT, Savage JE, Johnson EC, Huang Y, Edwards AC, Hottenga JJ, Grotzinger AD, Gustavson DE, Jennings MV, Anokhin A, Dick DM, Edenberg HJ, Kramer JR, Lai D, Meyers JL, Pandey AK, Harden KP, Nivard MG, de Geus EJC, Boomsma DI, Agrawal A, Davis LK, Clarke T-K, Palmer AA, Sanchez-Roige S (2022b) Item-level genome-wide Association study of the Alcohol Use Disorders Identification Test in Three Population-based cohorts. Am J Psychiatry 179:58–70. https://doi.org/10.1176/appi.ajp.2020.20091390
Marees AT, de Kluiver H, Stringer S, Vorspan F, Curis E, Marie-Claire C, Derks EM (2018) A tutorial on conducting genome-wide association studies: quality control and statistical analysis. Int J Methods Psychiatr Res 27:e1608. https://doi.org/10.1002/mpr.1608
McCarthy S, Das S, Kretzschmar W et al (2016) A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48:1279–1283. https://doi.org/10.1038/ng.3643
McVean GA et al (2012) Altshuler (Co-Chair) DM, Durbin (Co-Chair) RM, An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. https://doi.org/10.1038/nature11632
Meyers JL, Salvatore JE, Vuoksimaa E, Korhonen T, Pulkkinen L, Rose RJ, Kaprio J, Dick DM (2014) Genetic influences on alcohol use behaviors have diverging developmental trajectories: a prospective study among male and female twins. Alcohol Clin Exp Res 38:2869–2877. https://doi.org/10.1111/acer.12560
Nakagawa S, Schielzeth H (2013) A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol 4:133–142. https://doi.org/10.1111/j.2041-210x.2012.00261.x
Neale Lab (2018) UK Biobank GWAS Results. http://www.nealelab.is/uk-biobank
Northstone K, Lewcock M, Groom A, Boyd A, Macleod J, Timpson N, Wells N (2019) The Avon Longitudinal Study of parents and children (ALSPAC): an update on the enrolled sample of index children in 2019. Wellcome Open Res 4:51. https://doi.org/10.12688/wellcomeopenres.15132.1
Okbay A, Wu Y, Wang N, Jayashankar H, Bennett M, Nehzati SM, Sidorenko J, Kweon H, Goldman G, Gjorgjieva T, Jiang Y, Hicks B, Tian C, Hinds DA, Ahlskog R, Magnusson PKE, Oskarsson S, Hayward C, Campbell A, Porteous DJ, Freese J, Herd P, Watson C, Jala J, Conley D, Koellinger PD, Johannesson M, Laibson D, Meyer MN, Lee JJ, Kong A, Yengo L, Cesarini D, Turley P, Visscher PM, Beauchamp JP, Benjamin DJ, Young AI (2022) Polygenic prediction of educational attainment within and between families from genome-wide association analyses in 3 million individuals. Nat Genet 54:437–449. https://doi.org/10.1038/s41588-022-01016-z
Pagan JL, Rose RJ, Viken RJ, Pulkkinen L, Kaprio J, Dick DM (2006) Genetic and Environmental Influences on Stages of Alcohol Use Across Adolescence and into Young Adulthood. Behav Genet 36:483–497. https://doi.org/10.1007/s10519-006-9062-y
Paternoster R, Brame R, Mazerolle P, Piquero A (1998) Using the correct statistical test for the Equality of Regression coefficients. Criminology 36:859–866. https://doi.org/10.1111/j.1745-9125.1998.tb01268.x
Paternoster L, Howe LD, Tilling K, Weedon MN, Freathy RM, Frayling TM, Kemp JP, Smith GD, Timpson NJ, Ring SM, Evans DM, Lawlor DA (2011) Adult height variants affect birth length and growth rate in children. Hum Mol Genet 20:4069–4075. https://doi.org/10.1093/hmg/ddr309
Peterson RE, Kuchenbaecker K, Walters RK, Chen C-Y, Popejoy AB, Periyasamy S, Lam M, Iyegbe C, Strawbridge RJ, Brick L, Carey CE, Martin AR, Meyers JL, Su J, Chen J, Edwards AC, Kalungi A, Koen N, Majara L, Schwarz E, Smoller JW, Stahl EA, Sullivan PF, Vassos E, Mowry B, Prieto ML, Cuellar-Barboza A, Bigdeli TB, Edenberg HJ, Huang H, Duncan LE (2019) Genome-wide Association studies in Ancestrally diverse populations: opportunities, methods, pitfalls, and recommendations. Cell 0. https://doi.org/10.1016/j.cell.2019.08.051
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. https://doi.org/10.1086/519795
Purves KL, Coleman JRI, Meier SM, Rayner C, Davis KAS, Cheesman R, Bækvad-Hansen M, Børglum AD, Wan Cho S, Jürgen Deckert J, Gaspar HA, Bybjerg-Grauholm J, Hettema JM, Hotopf M, Hougaard D, Hübel C, Kan C, McIntosh AM, Mors O, Bo Mortensen P, Nordentoft M, Werge T, Nicodemus KK, Mattheisen M, Breen G, Eley TC (2020) A major role for common genetic variation in anxiety disorders. Mol Psychiatry 25:3292–3303. https://doi.org/10.1038/s41380-019-0559-1
R Core Team (2017) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Risch N, Merikangas K (1996) The future of genetic studies of complex human Diseases. Science 273:1516–1517. https://doi.org/10.1126/science.273.5281.1516
Rose RJ, Dick DM, Viken RJ, Kaprio J (2001) Gene-environment interaction in patterns of adolescent drinking: regional residency moderates longitudinal influences on alcohol use. Alcohol Clin Exp Res 25:637–643
Rosseel Y (2012) Lavaan: an R Package for Structural equation modeling. J Stat Softw 48:1–36. https://doi.org/10.18637/jss.v048.i02
Sakai JT, Stallings MC, Crowley TJ, Gelhorn HL, McQueen MB, Ehringer MA (2010) Test of association between GABRA2 (SNP rs279871) and adolescent conduct/alcohol Use Disorders utilizing a sample of clinic referred youth with serious substance and conduct problems, controls and available first degree relatives. Drug Alcohol Depend 106:199–203. https://doi.org/10.1016/j.drugalcdep.2009.08.015
Salvatore JE, Thomas NS, Cho SB, Adkins A, Kendler KS, Dick DM (2016) The role of romantic relationship status in pathways of risk for emerging adult alcohol use. Psychol Addict Behav 30:335–344. https://doi.org/10.1037/adb0000145
Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, Adams MJ, Howard DM, Edenberg HJ, Davies G, Crist RC, Deary IJ, McIntosh AM, Clarke T-K (2019) Genome-wide association study meta-analysis of the Alcohol Use Disorder Identification Test (AUDIT) in two population-based cohorts. Am J Psychiatry 176:107–118. https://doi.org/10.1176/appi.ajp.2018.18040369
Saunders GRB, Wang X, Chen F et al (2022) Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 1–7. https://doi.org/10.1038/s41586-022-05477-4
Thomas NS, Adkins A, Aliev F, Edwards AC, Webb BT, Tiarsmith EC, Kendler KS, Dick DM, Chartier KG (2018) Alcohol metabolizing polygenic risk for Alcohol Consumption in European American College Students. J Stud Alcohol Drugs 79:627–634. https://doi.org/10.15288/jsad.2018.79.627
Verhulst B, Neale MC, Kendler KS (2015) The heritability of Alcohol Use Disorders: a meta-analysis of twin and adoption studies. Psychol Med 45:1061–1072. https://doi.org/10.1017/S0033291714002165
Viken RJ, Kaprio J, Koskenvuo M, Rose RJ (1999) Longitudinal analyses of the determinants of drinking and of drinking to intoxication in adolescent twins. Behav Genet 29:455–461. https://doi.org/10.1023/a:1021631122461
Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J (2017) 10 years of GWAS Discovery: Biology, function, and translation. Am J Hum Genet 101:5–22. https://doi.org/10.1016/j.ajhg.2017.06.005
Wagenaar AC, Toomey TL (2015) Effects of minimum drinking age laws: review and analyses of the literature from 1960 to 2000. https://doi.org/10.15288/jsas.2002.s14.206. Journal of Studies on Alcohol, Supplement
Walters RK, Polimanti R, Johnson EC et al (2018) Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nat Neurosci 21:1656–1669. https://doi.org/10.1038/s41593-018-0275-1
Willer CJ, Li Y, Abecasis GR (2010) METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26:2190–2191. https://doi.org/10.1093/bioinformatics/btq340
World Health Organization (2018) Global Status Report on Alcohol and Health 2018. World Health Organization, Geneva, Switzerland
Yang J, Lee SH, Goddard ME, Visscher PM (2011) GCTA: A Tool for Genome-wide Complex Trait Analysis. Am J Hum Genet 88:76–82. https://doi.org/10.1016/j.ajhg.2010.11.011
Acknowledgements
COGA: The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, T. Foroud; Scientific Director, A. Agrawal; Translational Director, D. Dick, includes ten different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, T. Foroud, Y. Liu, M.H. Plawecki); University of Iowa Carver College of Medicine (S. Kuperman, J. Kramer); SUNY Downstate Health Sciences University (B. Porjesz, J. Meyers, C. Kamarajan, A. Pandey); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, D. Dick, R. Hart, J. Salvatore); The Children’s Hospital of Philadelphia, University of Pennsylvania (L. Almasy); Icahn School of Medicine at Mount Sinai (A. Goate, P. Slesinger); and Howard University (D. Scott). Other COGA collaborators include: L. Bauer (University of Connecticut); J. Nurnberger Jr., L. Wetherill, X., Xuei, D. Lai, S. O’Connor, (Indiana University); G. Chan (University of Iowa; University of Connecticut); D.B. Chorlian, J. Zhang, P. Barr, S. Kinreich, G. Pandey (SUNY Downstate); N. Mullins (Icahn School of Medicine at Mount Sinai); A. Anokhin, S. Hartz, E. Johnson, V. McCutcheon, S. Saccone (Washington University); J. Moore, F. Aliev, Z. Pang, S. Kuo (Rutgers University); A. Merikangas (The Children’s Hospital of Philadelphia and University of Pennsylvania); H. Chin and A. Parsian are the NIAAA Staff Collaborators. We continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting- Kai Li, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
ALSPAC: We are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. The UK Medical Research Council and Wellcome (Grant ref: 217065/Z/19/Z) and the University of Bristol provide core support for ALSPAC. This publication is the work of the authors and will serve as guarantors for the contents of this paper. A comprehensive list of grants funding is available on the ALSPAC website. This research was specifically funded by NIH AA018333, MRC G0800612/86812, Wellcome Trust and MRC (Core) 076467/Z/05/, NIH 5R01AA018333-05, Wellcome Trust and MRC 092731. GWAS data was generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe.
Add Health: Add Health is directed by Robert A. Hummer and funded by the National Institute on Aging cooperative agreements U01 AG071448 (Hummer) and U01AG071450 (Aiello and Hummer) at the University of North Carolina at Chapel Hill. Waves I-V data are from the Add Health Program Project, grant P01 HD31921 (Harris) from Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), with cooperative funding from 23 other federal agencies and foundations. Add Health was designed by J. Richard Udry, Peter S. Bearman, and Kathleen Mullan Harris at the University of North Carolina at Chapel Hill.
High Performance Computing resources provided by the High Performance Research Computing (HPRC) core facility at Virginia Commonwealth University (https://hprc.vcu.edu) and HPC resources hosted by the Virginia Institute for Psychiatric and Behavioral Genetics were used for conducting the research reported in this work.
Funding
This work was supported by the National Institutes of Health (NIH) Grant F31AA029620 (PI: Thomas) from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and T32DA015035 (APM) from the National Institute on Drug Abuse (NIDA).
COGA: The Collaborative Study on the Genetics of Alcoholism (COGA) is supported by NIH Grant U10AA008401 (PI: Porjesz).
ALSPAC: A comprehensive list of grants funding is available on the ALSPAC website. This research was specifically funded by NIH AA018333, MRC G0800612/86812, Wellcome Trust and MRC (Core) 076467/Z/05/, NIH 5R01AA018333-05, Wellcome Trust and MRC 092731.
AddHealth: Add Health is funded by the National Institute on Aging cooperative agreements U01 AG071448 (Hummer) and U01AG071450 (Aiello and Hummer) at the University of North Carolina at Chapel Hill. Waves I-V data are from the Add Health Program Project, grant P01 HD31921 (Harris) from Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), with cooperative funding from 23 other federal agencies and foundations.
Author information
Authors and Affiliations
Contributions
Nathaniel S. Thomas: conceived of the study, conducted statistical analyses, and wrote the manuscript. Nathan A. Gillespie assisted with the design and implementation of the study and provided editorial feedback on the whole manuscript. Grace Chan, Howard J. Edenberg, Chella Kamarajan, Sally I-Chun Kuo, Alex P. Miller, John I. Nurnberger Jr., and Jay Tischfield provided editorial feedback on the whole manuscript. Jessica E. Salvatore and Danielle M. Dick supervised the design and implementation of the study and provided editorial feedback on the whole manuscript. All authors contributed to and have approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of Interest/Competing Interests
Nathaniel S. Thomas, Nathan A. Gillespie, Grace Chan, Howard J. Edenberg, Chella Kamarajan, Sally I-Chun Kuo, Alex P. Miller, John I. Nurnberger Jr., Jay Tischfield, Danielle M. Dick, and Jessica E. Salvatore declare that they have no conflicts of interest.
Ethics Approval
Secondary analysis of these data was determined to be qualified for exemption by the Institutional Review Board at Virginia Commonwealth University (HM20024009) according to 45 CFR 46 under exempt category 4 (ii). Ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees (http://www.bristol.ac.uk/alspac/researchers/research-ethics/).
Consent to Participate
Written consent was obtained from all participants by each respective study.
Consent for Publication
NA.
Additional information
Edited by Tinca Polderman
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Danielle M. Dick and Jessica E. Salvatore jointly supervised this work.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Thomas, N.S., Gillespie, N.A., Chan, G. et al. A Developmentally-Informative Genome-wide Association Study of Alcohol Use Frequency. Behav Genet 54, 151–168 (2024). https://doi.org/10.1007/s10519-023-10170-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-023-10170-x