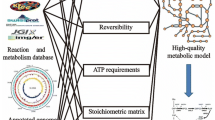
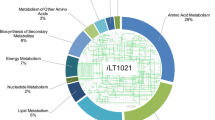
Abstract
Genome-scale metabolic reconstructions are routinely used for the analysis and design of metabolic engineering strategies for production of primary metabolites. The use of such reconstructions for metabolic engineering of antibiotic production is not common due to the lack of simple design algorithms in the absence of a cellular growth objective function. Here, we present the metabolic network reconstruction for the erythromycin producer Saccharopolyspora erythraea NRRL23338. The model was manually curated for primary and secondary metabolism pathways and consists of 1,482 reactions (2,075 genes) and 1,646 metabolites. As part of the model validation, we explored the potential benefits of supplying amino acids and identified five amino acids “compatible” with erythromycin production, whereby if glucose is supplemented with this amino acid on a carbon mole basis, the in silico model predicts that high erythromycin yield is possible without lowering biomass yield. Increased erythromycin titre was confirmed for four of the five amino acids, namely valine, isoleucine, threonine and proline. In bioreactor experiments, supplementation with 2.5 % carbon mole of valine increased the growth rate by 20 % and simultaneously the erythromycin yield on biomass by 50 %. The model presented here can be used as a framework for the future integration of high-throughput biological data sets in S. erythraea and ultimately to realise strain designs capable of increasing erythromycin production closer to the theoretical yield.



Similar content being viewed by others
References
Alam MT, Merlo M, Consortium TS, Hodgson D, Wellington E, Takano E, Breitling R (2010) Metabolic modeling and analysis of the metabolic switch in Streptomyces coelicolor. BMC Genomics 11(1):202
Alam MT, Medema MH, Takano E, Breitling R (2011) Comparative genome-scale metabolic modeling of actinomycetes: the topology of essential core metabolism. FEBS Lett 585(14):2389–2394
Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R, Formsma K, Gerdes S, Glass E, Kubal M, Meyer F, Olsen G, Olson R, Osterman A, Overbeek R, McNeil L, Paarmann D, Paczian T, Parrello B, Pusch G, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9(1):75
Bahal N, Nahata MC (1992) The new macrolide antibiotics: azithromycin, clarithromycin, dirithromycin, and roxithromycin. Ann Pharmacother 26(1):46–55
Barona-Gomez F, Hodgson DA (2003) Occurrence of a putative ancient-like isomerase involved in histidine and tryptophan biosynthesis. EMBO Rep 4(3):296–300
Barona-Gomez F, Cruz-Morales P, Noda-García L (2012) What can genome-scale metabolic network reconstructions do for prokaryotic systematics? Antonie Van Leeuwenhoek 101(1):35–43
Beste D, Hooper T, Stewart G, Bonde B, Avignone-Rossa C, Bushell M, Wheeler P, Klamt S, Kierzek A, McFadden J (2007) GSMN-TB: a web-based genome-scale network model of Mycobacterium tuberculosis metabolism. Genome Biol 8 (5):R89
Borodina I, Krabben P, Nielsen J (2005) Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res 15(6):820–829
Bott M, Niebisch A (2003) The respiratory chain of Corynebacterium glutamicum. J Biotechnol 104(1–3):129–153
Burgard AP, Pharkya P, Maranas CD (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 84(6):647–657
Chen WY, Marcellin E, Hung J, Nielsen LK (2009) Hyaluronan molecular weight s controlled by UDP-N-acetylglucosamine concentration in Streptococcus zooepidemicus. J Biol Chem 284(27):18007–18014
Chindelevitch L, Stanley S, Hung D, Regev A, Berger B (2012) MetaMerge: scaling up genome-scale metabolic reconstructions with application to Mycobacterium tuberculosis. Genome Biol 13 (1):r8
Cortés J, Velasco J, Foster G, Blackaby AP, Rudd BAM, Wilkinson B (2002) Identification and cloning of a type III polyketide synthase required for diffusible pigment biosynthesis in Saccharopolyspora erythraea. Mol Microbiol 44(5):1213–1224
de Oliveira Dal’Molin CG, Quek L-E, Palfreyman RW, Brumbley SM, Nielsen LK (2009) AraGEM, a genome-scale reconstruction of the primary metabolic network in Arabidopsis. Plant Physiol 152(2):579–589
Edwards JS, Palsson BO (1999) Systems properties of the Haemophilus influenzaeRd metabolic genotype. J Biol Chem 274(25):17410–17416
Edwards JS, Palsson BO (2000) The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Nat Acad Sci USA 97(10):5528–5533
El-Enshasy HA, Mohamed NA, Farid MA, El-Diwany AI (2008) Improvement of erythromycin production by Saccharopolyspora erythraea in molasses based medium through cultivation medium optimization. Bioresour Technol 99(10):4263–4268
Hamedi J, Malekzadeh F, Saghafi-nia AE (2004) Enhancing of erythromycin production by Saccharopolyspora erythraea with common and uncommon oils. J Ind Microbiol Biotechnol 31(10):447–456
Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL (2010) High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotech 28(9):977–982
Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H, Dairi T (2008) An alternative menaquinone biosynthetic pathway operating in microorganisms. Science 321(5896):1670–1673
Ingraham JL, Maaloe O, Neidhart FC (1983) Growth of the bacterial cell. Sinauer Associates, Massachusetts
Jamshidi N, Palsson B (2007) Investigating the metabolic capabilities of Mycobacterium tuberculosis H37Rv using the in silico strain iNJ661 and proposing alternative drug targets. BMC Syst Biol 1(1):26
Jung W, Yoo Y, Park J, Park S, Han A, Ban Y, Kim E, Kim E, Yoon Y (2011) A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicui ATCC 29253. Appl Microbiol Biotechnol 91(5):1389–1397
Kim TY, Kim HU, Lee SY (2010) Metabolite-centric approaches for the discovery of antibacterials using genome-scale metabolic networks. Metab Eng 12(2):105–111
Kjeldsen KR, Nielsen J (2009) In silico genome-scale reconstruction and validation of the corynebacterium glutamicum metabolic network. Biotechnol Bioeng 102(2):583–597
Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H (2008) Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proceedings of the National Academy of Sciences
Labeda DP (1987) Transfer of the type strain of Streptomyces erythreus (Waksman 1923) Waksman and Henrici 1948 to the genus Saccharopolyspora Lacey and Goodfellow 1975 as Saccharopolyspora erythraea sp. nov., and designation of a neotype strain for Streptomyces erythraeus. Int J Syst Bacteriol 37(1):19–22
Lazos O, Tosin M, Slusarczyk AL, Boakes S, Cortes J, Sidebottom PJ, Leadlay PF (2010) Biosynthesis of the putative siderophore erythrochelin requires unprecedented crosstalk between separate nonribosomal peptide gene clusters. Chem Biol 17(2):160–173
Medema MH, Trefzer A, Kovalchuk A, van den Berg M, Muller U, Heijne W, Wu L, Alam MT, Ronning CM, Nierman WC, Bovenberg RAL, Breitling R, Takano E (2010) The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol Evol 2:212224
Medema MH, Alam MT, Heijne WHM, van den Berg MA, Müller U, Trefzer A, Bovenberg RAL, Breitling R, Takano E (2011) Genome-wide gene expression changes in an industrial clavulanic acid overproduction strain of Streptomyces clavuligerus. Microb Biotechnol 4(2):300–305
Mormann S, Lomker A, Ruckert C, Gaigalat L, Tauch A, Puhler A, Kalinowski J (2006) Random mutagenesis in Corynebacterium glutamicum ATCC 13032 using an IS6100-based transposon vector identified the last unknown gene in the histidine biosynthesis pathway. BMC Genomics 7(1):205
Nett M, Ikeda H, Moore BS (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26(11):1362–1384
Nieselt K, Battke F, Herbig A, Bruheim P, Wentzel A, Jakobsen O, Sletta H, Alam M, Merlo M, Moore J, Omara W, Morrissey E, Juarez-Hermosillo M, Rodriguez-Garcia A, Nentwich M, Thomas L, Iqbal M, Legaie R, Gaze W, Challis G, Jansen R, Dijkhuizen L, Rand D, Wild D, Bonin M, Reuther J, Wohlleben W, Smith M, Burroughs N, Martin J, Hodgson D, Takano E, Breitling R, Ellingsen T, Wellington E (2010) The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics 11(1):10
Oberhardt MA, Palsson BO, Papin JA (2009) Applications of genome-scale metabolic reconstructions. Mol Syst Biol 5:320
Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF (2007) Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotech 25(4):447–453
Orth JD, Thiele I, Palsson BO (2010) What is flux balance analysis? Nat Biotech 28(3):245–248
Park JH, Lee KH, Kim TY, Lee SY (2007) Metabolic engineering of Escherichia coli for the production of l-valine based on transcriptome analysis and in silico gene knockout simulation. Proc Nat Acad Sci 104(19):7797–7802
Peano C, Bicciato S, Corti G, Ferrari F, Rizzi E, Bonnal R, Bordoni R, Albertini A, Bernardi L, Donadio S, De Bellis G (2007) Complete gene expression profiling of Saccharopolyspora erythraea using GeneChip DNA microarrays. Microb Cell Fact 6(1):37
Price ND, Reed JL, Palsson BO (2004) Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Micro 2(11):886–897
Quek L-E, Nielsen L (2008) On the reconstruction of the Mus musculus genome-scale metabolic network model. Genome Inform 21:11
Reeves A, Brikun I, Cernota W, Leach B, Gonzalez M, Weber J (2006) Effects of methylmalonyl-CoA mutase gene knockouts on erythromycin production in carbohydrate-based and oil-based fermentations of Saccharopolyspora erythraea. J Ind Microbiol Biotechnol 33(7):600–609
Reeves AR, Brikun IA, Cernota WH, Leach BI, Gonzalez MC, Mark Weber J (2007) Engineering of the methylmalonyl-CoA metabolite node of Saccharopolyspora erythraea for increased erythromycin production. Metab Eng 9(3):293–303
Robbel L, Knappe TA, Linne U, Xie X, Marahiel MA (2009) Erythrochelin—a hydroxamate-type siderophore predicted from the genome of Saccharopolyspora erythraea. FEBS J 277(3):663–676
Robbel L, Helmetag V, Knappe TA, Marahiel MA (2011) Consecutive enzymatic modification of ornithine generates the hydroxamate moieties of the siderophore erythrochelin. Biochemistry 50(27):6073–6080
Sandoval-Calderon M, Geiger O, Guan Z, Barona-Gomez F, Sohlenkamp C (2009) A Eukaryote-like cardiolipin synthase is present in Streptomyces coelicolor and in most actinobacteria. J Biol Chem 284(26):17383–17390
Schilling CH, Covert MW, Famili I, Church GM, Edwards JS, Palsson BO (2002) Genome-scale metabolic model of Helicobacter pylori 26695. J Bacteriol 184(16):4582–4593
Shinfuku Y, Sorpitiporn N, Sono M, Furusawa C, Hirasawa T, Shimizu H (2009) Development and experimental verification of a genome-scale metabolic model for Corynebacterium glutamicum. Microb Cell Fact 8(1):43
Tanaka Y, Komatsu M, Okamoto S, Tokuyama S, Kaji A, Ikeda H, Ochi K (2009) Antibiotic overproduction by rpsL and rsmG mutants of various actinomycetes. Appl Environ Microbiol 75(14):4919–4922
Tang L, Zhang YX, Hutchinson CR (1994) Amino acid catabolism and antibiotic synthesis: valine is a source of precursors for macrolide biosynthesis in Streptomyces ambofaciens and Streptomyces fradiae. J Bacteriol 176(19):6107–6119
Weber JM, Leung JO, Maine GT, Potenz RH, Paulus TJ, DeWitt JP (1990) Organization of a cluster of erythromycin genes in Saccharopolyspora erythraea. J Bacteriol 172(5):2372–2383
Zou X, Hang H-f, Chu J, Zhuang Y-p, Zhang S-l (2009) Oxygen uptake rate optimization with nitrogen regulation for erythromycin production and scale-up from 50 L to 372 m3 scale. Bioresour Technol 100(3):1406–1412
Acknowledgments
We gratefully acknowledge the financial support of the Mexican Council for Science and Technology (CONACyT) and the Australian Institute for Bioengineering and Nanotechnology (AIBN). We further would like to thank Michael Wang for support with HPLC analysis.
Conflict of interest
The authors have declared no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Licona-Cassani, C., Marcellin, E., Quek, LE. et al. Reconstruction of the Saccharopolyspora erythraea genome-scale model and its use for enhancing erythromycin production. Antonie van Leeuwenhoek 102, 493–502 (2012). https://doi.org/10.1007/s10482-012-9783-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-012-9783-2