Abstract
Background
Magnetic Resonance Imaging (MRI) is a highly demanded medical imaging system due to high resolution, large volumetric coverage, and ability to capture the dynamic and functional information of body organs e.g. cardiac MRI is employed to assess cardiac structure and evaluate blood flow dynamics through the cardiac valves. Long scan time is the main drawback of MRI, which makes it difficult for the patients to remain still during the scanning process.
Objective
By collecting fewer measurements, MRI scan time can be shortened, but this undersampling causes aliasing artifacts in the reconstructed images. Advanced image reconstruction algorithms have been used in literature to overcome these undersampling artifacts. These algorithms are computationally expensive and require a long time for reconstruction which makes them infeasible for real-time clinical applications e.g. cardiac MRI. However, exploiting the inherent parallelism in these algorithms can help to reduce their computation time.
Methods
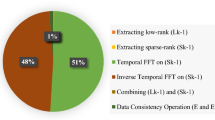
Low-rank plus sparse (L+S) matrix decomposition model is a technique used in literature to reconstruct the highly undersampled dynamic MRI (dMRI) data at the expense of long reconstruction time. In this paper, Compressed Singular Value Decomposition (cSVD) model is used in L+S decomposition model (instead of conventional SVD) to reduce the reconstruction time. The results provide improved quality of the reconstructed images. Furthermore, it has been observed that cSVD and other parts of the L+S model possess highly parallel operations; therefore, a customized GPU based parallel architecture of the modified L+S model has been presented to further reduce the reconstruction time.
Results
Four cardiac MRI datasets (three different cardiac perfusion acquired from different patients and one cardiac cine data), each with different acceleration factors of 2, 6 and 8 are used for experiments in this paper. Experimental results demonstrate that using the proposed parallel architecture for the reconstruction of cardiac perfusion data provides a speed-up factor up to 19.15× (with memory latency) and 70.55× (without memory latency) in comparison to the conventional CPU reconstruction with no compromise on image quality.
Conclusion
The proposed method is well-suited for real-time clinical applications, offering a substantial reduction in reconstruction time.












Similar content being viewed by others
Data availability
The research article utilizes data gathered from various sources, as appropriately cited within the document. Additionally, a dedicated repository has been established for citing references related to this research article as: https://figshare.com/s/b28d3066076439ae680d
Change history
31 December 2023
Few spacing errors have been updated in the abstract and text has been changed from “60 temporal frame” to “60 temporal frames”.
References
McRobbie DW, Moore EA (2007) MRI, from picture to proton. Clin Radiol 62:1233
Zaitsev M, Maclaren J, Herbst M (2015) Motion artifacts in MRI: a complex problem with many partial solutions. J Magn Reson Imaging 42(4):887–901
Ohliger MA, Grant AK, Sodickson DK (2003) Ultimate intrinsic signal-to-noise ratio for parallel MRI: electromagnetic field considerations. Magn Reson Med 50(5):1018–1030
Blaimer M, Breuer F, Mueller M, Heidemann RM, Griswold MA, Jakob PM (2004) SMASH, SENSE, PILS, GRAPPA: how to choose the optimal method. Top Magn Reson Imaging 15(4):223–236
Lustig M, Donoho D, Pauly JM (2007) Sparse MRI: the application of compressed sensing for rapid MR imaging. Magn Reson Med 58(6):1182–1195
Chandarana H et al (2013) Free-breathing contrast-enhanced multiphase MRI of the liver using a combination of compressed sensing, parallel imaging, and golden-angle radial sampling. Investig Radiol 48(1):10
King K, Xu D, Brau A, Lai P, Beatty PJ, Marinelli L (2010) A new combination of compressed sensing and data driven parallel imaging. In: Proceedings of the 18th scientific meeting, International Society for Magnetic Resonance in Medicine, Stockholm
Otazo R, Feng L, Chandarana H, Block T, Axel L, Sodickson DK (2012) Combination of compressed sensing and parallel imaging for highly-accelerated dynamic MRI. In: 2012 9th IEEE International Symposium on Biomedical Imaging (Isbi), pp 980–983
Otazo R, Kim D, Axel L, Sodickson DK (2011) Combination of compressed sensing and parallel imaging with respiratory motion correction for highly-accelerated cardiac perfusion MRI. J Cardiovasc Magn Reson 13(1):1–2
Otazo R, Candes E, Sodickson DK (2015) Low-rank plus sparse matrix decomposition for accelerated dynamic MRI with separation of background and dynamic components. Magn Reson Med 73(3):1125–1136
Huang W et al (2021) Deep low-rank plus sparse network for dynamic MR imaging. Med Image Anal 73:102190
Lebrun M, Leclaire A (2012) An implementation and detailed analysis of the K-SVD image denoising algorithm. Image Process Line 2:96–133
Wei J-J, Chang C-J, Chou N-K, Jan G-J (2001) ECG data compression using truncated singular value decomposition. IEEE Trans Inf Technol Biomed 5(4):290–299
Golub G, Kahan W (1965) Calculating the singular values and pseudo-inverse of a matrix. J Soc Ind Appl Math Ser B Numer Anal 2(2):205–224
Zhang L, Dong W, Zhang D, Shi G (2010) Two-stage image denoising by principal component analysis with local pixel grouping. Pattern Recogn 43(4):1531–1549
Zhang T, Pauly JM, Vasanawala SS, Lustig M (2013) Coil compression for accelerated imaging with Cartesian sampling. Magn Reson Med 69(2):571–582
Qazi SA, Saeed A, Nasir S, Omer H (2017) Singular value decomposition using Jacobi algorithm in pMRI and CS. Appl Magn Reson 48(5):461–471
Demmel J, Veselić K (1992) Jacobi’s method is more accurate than QR. SIAM J Matrix Anal Appl 13(4):1204–1245
Benjamin Erichson N, Brunton SL, Nathan Kutz J (2017) Compressed singular value decomposition for image and video processing. In: Proceedings of the IEEE International Conference on Computer Vision Workshops, pp 1880–1888
Mahoney MW (2011) Randomized algorithms for matrices and data. arXiv preprint arXiv:1104.5557
Stone SS, Haldar JP, Tsao SC, Sutton B, Liang Z-P (2008) Accelerating advanced MRI reconstructions on GPUs. J Parallel Distrib Comput 68(10):1307–1318
Kirk DB, Wen-Mei WH (2016) Programming massively parallel processors: a hands-on approach. Morgan kaufmann
Khalil MA, Ashfaq A, Shahzad H, Qazi SA, Omer H (2021) GPU based parallel framework for receiver coil sensitivity estimation in SENSE reconstruction. Magn Reson Imaging 80:58–70
Qazi SA, Nasir S, Saeed A, Omer H (2018) Optimizing image reconstruction in SENSE using GPU. Appl Magn Reson 49:151–164
Shahzad H, Sadaqat M, Hassan B, Abbasi W, Omer H (2016) Parallel MRI reconstruction algorithm implementation on GPU. Appl Magn Reson 47(1):53–61
Ullah I, Nisar H, Raza H, Qasim M, Inam O, Omer H (2018) QR-decomposition based SENSE reconstruction using parallel architecture. Comput Biol Med 95:1–12
Qazi SA, Tariq F, Ullah I, Omer H (2021) Parallel implementation of L+ S signal recovery in dynamic MRI. Magn Reson Mater Phys Biol Med 34(2):297–307
Shafique M, Qazi SA, Ullah I, Omer H (2022) Compressed SVD for L+S matrix decomposition model to reconstruct undersampled dynamic MRI. In: Joint Annual Meeting ISMRM-ESMRMB & ISMRT 31st Annual Meeting
Cai J-F, Candès EJ, Shen Z (2010) A singular value thresholding algorithm for matrix completion. SIAM J Optim 20(4):1956–1982
Candes EJ, Romberg JK, Tao T (2006) Stable signal recovery from incomplete and inaccurate measurements. Commun Pure Appl Math 59(8):1207–1223
Sanders J, Kandrot E (2010) CUDA by example: an introduction to general-purpose GPU programming. Addison-Wesley Professional
Gou C, Gaydadjiev GN (2013) Addressing GPU on-chip shared memory bank conflicts using elastic pipeline. Int J Parallel Prog 41(3):400–429
Andreopoulos A, Tsotsos JK (2008) Efficient and generalizable statistical models of shape and appearance for analysis of cardiac MRI. Med Image Anal 12(3):335–357
Walsh DO, Gmitro AF, Marcellin MW (2000) Adaptive reconstruction of phased array MR imagery. Magn Reson Med 43(5):682–690
Liu S, Schniter P, Ahmad R (2023) MRI recovery with self-calibrated denoisers without fully-sampled data. arXiv preprint arXiv:2304.12890
Uecker M et al (2014) ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magn Reson Med 71(3):990–1001
Ji JX, Son JB, Rane SD (2007) PULSAR: a Matlab toolbox for parallel magnetic resonance imaging using array coils and multiple channel receivers. Concepts Magn Reson Part B Magn Reson Eng Educ J 31(1):24–36
Chow LS, Rajagopal H, Paramesran R, Initiative ASDN (2016) Correlation between subjective and objective assessment of magnetic resonance (MR) images. Magn Reson Imaging 34(6):820–831
Omer H, Dickinson R (2010) A graphical generalized implementation of SENSE reconstruction using Matlab. Concepts Magn Reson Part A 36(3):178–186
Golub GH (1996) Cf vanloan, matrix computations. Johns Hopkins 113(10):23–36
Mahoney MW (2011) Randomized algorithms for matrices and data. Found Trends Mach Learn 3(2):123–224
Ravishankar S, Bresler Y (2010) MR image reconstruction from highly undersampled k-space data by dictionary learning. IEEE Trans Med Imaging 30(5):1028–1041
Chen C et al (2019) Sparsity adaptive reconstruction for highly accelerated cardiac MRI. Magn Reson Med 81(6):3875–3887
Khare K, Hardy CJ, King KF, Turski PA, Marinelli L (2012) Accelerated MR imaging using compressive sensing with no free parameters. Magn Reson Med 68(5):1450–1457
Weller DS, Ramani S, Nielsen JF, Fessler JA (2014) Monte Carlo SURE-based parameter selection for parallel magnetic resonance imaging reconstruction. Magn Reson Med 71(5):1760–1770
Dar SUH, Özbey M, Çatlı AB, Çukur T (2020) A transfer-learning approach for accelerated MRI using deep neural networks. Magn Reson Med 84(2):663–685
Elmas G et al (2022) Federated learning of generative image priors for MRI reconstruction. IEEE Trans Med Imaging 42:1996
Güngör A et al (2023) Adaptive diffusion priors for accelerated MRI reconstruction. Med Image Anal
Korkmaz Y, Dar SU, Yurt M, Özbey M, Cukur T (2022) Unsupervised MRI reconstruction via zero-shot learned adversarial transformers. IEEE Trans Med Imaging 41(7):1747–1763
Yosinski J, Clune J, Bengio Y, Lipson H (2014) How transferable are features in deep neural networks?. Adv Neural Inf Process Syst
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shafique, M., Qazi, S.A. & Omer, H. Compressed SVD-based L + S model to reconstruct undersampled dynamic MRI data using parallel architecture. Magn Reson Mater Phy (2023). https://doi.org/10.1007/s10334-023-01128-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10334-023-01128-5