Abstract
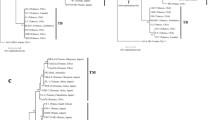
Three evolutionary lineages of the tomato wilt pathogen Fusarium oxysporum f. sp. lycopersici were found among a worldwide sample of isolates based on phylogenetic analysis of the ribosomal DNA intergenic spacer region. Each lineage consisted of isolates mainly belonging to a single or closely related vegetative compatibility group (VCG) and a single mating type (MAT). The first lineage (A1) was composed of isolates VCG 0031 and MAT1-1; the second (A2) included VCG 0030 and/or 0032 and MAT1-1; and the third (A3) included VCG 0033 and MAT1-2. Race 1 and race 2 isolates belonged to the A1 or A2 lineages, and race 3 belonged to A2 or A3 lineages, suggesting that there is no correlation between race and lineage. However, for the isolates from Japan, race 1 (with one exception), race 2, and race 3 isolates belonged to A2, A1, and A3 lineages, respectively. These results suggest that the races could have evolved independently in each lineage; and in Japan the present races were likely to have been introduced independently after they had evolved in other locations.
Similar content being viewed by others
References
LJ Alexander CM Tucker (1945) ArticleTitlePhysiologic specialization in the tomato wilt fungus Fusarium oxysporum f. lycopersici J Agric Res 70 303–313
DJ Appel TR Gordon (1996) ArticleTitleRelationships among pathogenic and nonpathogenic isolates of Fusarium oxysporum based on the partial sequence of the intergenic spacer region of the ribosomal DNA Mol Plant Microbe Interact 9 125–138 Occurrence Handle8820752
T Arie SK Christiansen OC Yoder BG Turgeon (1997) ArticleTitleEfficient cloning of ascomycete mating type genes by PCR amplification of the conserved MAT HMG box Fungal Genet Biol 21 118–130 Occurrence Handle10.1006/fgbi.1997.0961 Occurrence Handle9073486
T Arie S Gouthu S Shimazaki T Kamakura M Kimura M Inoue K Takio A Ozaki Y Yoneyama I Yamaguchi (1998) ArticleTitleImmunological detection of endopolygalacturonase secretion by Fusarium oxysporum in plant tissue and sequencing of its encoding gene Ann Phytopathol Soc Jpn 64 7–15
T Arie T Yoshida T Shimizu M Kawabe K Yoneyama I Yamaguchi (1999) ArticleTitleAssessment of Gibberella fujikuroi mating type by PCR Mycoscience 40 311–314
T Arie I Kaneko T Yoshida M Noguchi Y Nomura I Yamaguchi (2000) ArticleTitleMating-type genes from asexual phytopathogenic ascomycetes Fusarium oxysporum and Alternaria alternata Mol Plant Microbe Interact 13 1330–1339 Occurrence Handle11106025
MP Barve T Arie S Salimath FJ Muehlbauer TL Peever (2003) ArticleTitleCloning and characterization of mating type (MAT) locus from Ascochyta rabiei (telemorph: Didymella rabiei) and a MAT phylogeny of legunum-associated Ascochyta s Fungal Genet Biol 39 151–167 Occurrence Handle10.1016/S1087-1845(03)00015-X Occurrence Handle12781674
Booth C (1971) The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey, UK
G Cai LR Gale RW Schneider HC Kistler RM Davis KS Elias EM Miyao (2003) ArticleTitleOrigin of race 3 of Fusarium oxysporum f. sp. lycopersici at a single site in California Phytopathology 93 1014–1022
JC Correll CJR Klittich JF Leslie (1987) ArticleTitleNitrate nonutilizing mutants of Fusarium oxysporum and their use in vegetative compatibility tests Phytopathology 77 1640–1646
A Di Pietro MIG Roncero (1996) ArticleTitleEndopolygalacturonase from Fusarium oxysporum f. sp. lycopersici: purification, characterization, and production during infection of tomato plants Phytopathology 86 1324–1330
A Di Pietro MIG Roncero (1998) ArticleTitleCloning, expression, and role in pathogenicity of pg1 encoding the major extracellular endopolygalacturonase of the vascular wilt pathogen Fusarium oxysporum Mol Plant Microbe Interact 11 91–98 Occurrence Handle9450333
KS Elias RW Schneider (1991) ArticleTitleVegetative compatibility groups in Fusarium oxysporum f. sp. lycopersici Phytopathology 81 159–162
KS Elias RW Schneider (1992) ArticleTitleGenetic diversity within and among races and vegetative compatibility groups of Fusarium oxysporum f. sp. lycopersici as determined by isozyme analysis Phytopathology 82 1421–1427
KS Elias D Zamir T Lichtman-Pleban T Katan (1993) ArticleTitlePopulation structure of Fusarium oxysporum f. sp. lycopersici: restriction fragment length polymorphisms provide genetic evidence that vegetative compatibility group is an indicator of evolutionary origin Mol Plant Microbe Interact 6 565–572
J Felsenstein (1981) ArticleTitleEvolutionary tree from DNA sequences: a maximum likelihood approach J Mol Evol 39 783–791
Felsenstein J (1993) PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle
WM Fitch (1977) ArticleTitleOn the problem of discovering the most parsimonious tree Am Nat 111 223–257 Occurrence Handle10.1086/283157
LR Gale T Katan HC Kistler (2003) ArticleTitleThe probable center of origin of Fusarium oxysporum f. sp. lycopersici VCG 0033 Plant Dis 87 1433–1438
FI García-Maceira A Di Pietro MIG Roncero (2000) ArticleTitleCloning and distribution of pgx4 encoding an in planta expressed exopoly-galacturonase from Fusarium oxysporum Mol Plant Microbe Interact 13 359–365 Occurrence Handle10755298
E Gómez-Gómez MC Ruíz-Roldán A Di Pietro MI Roncero C Hera (2002) ArticleTitleRole in pathogenesis of two endo-β-1,4-xylanase genes from the vascular wilt fungus Fusarium oxysporum Fungal Genet Biol 35 213–333 Occurrence Handle10.1006/fgbi.2001.1318 Occurrence Handle11929211
TR Gordon RD Martyn (1997) ArticleTitleThe evolutionary biology of Fusarium oxysporum Annu Rev Phytopathol 35 111–128 Occurrence Handle10.1146/annurev.phyto.35.1.111 Occurrence Handle15012517
R Grattidge RG O’Brien (1982) ArticleTitleThe occurrence of a third race of Fusarium wilt of tomato in Queensland Plant Dis 66 165–166
Y Hosobuchi (1998) ArticleTitleFusarium oxysporum infecting fusarium wilt resistant tomato cultivars Ann Phytopathol Soc Jpn 64 434
MD Huertas-González MC Ruiz-Roldán FIG Maceria MIG Roncero A Di Pietro (1999) ArticleTitleCloning and characterization of pl1 encoding an in planta-secreted pectate lyase of Fusarium oxysporum Curr Genet 35 36–40 Occurrence Handle10.1007/s002940050430 Occurrence Handle10022947
T Katan (1999) ArticleTitleCurrent status of vegetative compatibility groups in Fusarium oxysporum Phytoparasitica 27 51–64
T Katan P Di Primo (1999) ArticleTitleCurrent status of vegetative compatibility groups in Fusarium oxysporum: supplement Phytoparasitica 27 273–277
T Katan D Zamir M Sarfatti J Katan (1991) ArticleTitleVegetative compatibility groups and subgroups in Fusarium oxysporum f. sp. radicis-lycopersici Phytopathology 81 255–262
M Kawabe K Mizutani T Yoshida T Teraoka K Yoneyama I Yamaguchi T Arie (2004) ArticleTitleCloning a pathogenicity-related gene, FPD1, in Fusarium oxysporum f. sp. lycopersici J Gen Plant Pathol 70 16–20 Occurrence Handle10.1007/s10327-003-0089-0
M Kimura (1980) ArticleTitleA simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences J Mol Evol 16 111–120 Occurrence Handle7463489
HC Kistler (1997) ArticleTitleGenetic diversity in the plant-pathogenic fungus Fusarium oxysporum Phytopathology 87 474–479
S Kumar K Tamura IB Jakobsen M Nei (2001) ArticleTitleMEGA2: molecular evolutionary genetics analysis software Bioinformatics 17 1244–1245 Occurrence Handle10.1093/bioinformatics/17.12.1244 Occurrence Handle11751241
S Kuninaga R Yokosawa (1992) ArticleTitleGenetic diversity of Fusarium oxysporum f. sp. lycopersici in restriction fragment length polymorphisms of mitochondrial DNA Trans Mycol Soc Jpn 33 449–459
JF Leslie (1993) ArticleTitleFungal vegetative compatibility Annu Rev Phytopathol 31 127–150
ML Marlatt JC Correll P Kaufmann (1996) ArticleTitleTwo genetically distinct populations of Fusarium oxysporum f. sp. lycopersici race 3 in the United States Plant Dis 80 1336–1342
T Masunaga H Shiomi H Komada (1998) ArticleTitleIdentification of race 3 of Fusarium oxysporum f. sp. lycopersici isolated from tomato in Fukuoka Prefecture Ann Phytopathol Soc Jpn 64 435
JJ Mes EA Weststeijn F Herlaar JJM Lambalk J Wijbrandi MA Haring BJC Cornelissen (1999) ArticleTitleBiological and molecular characterization of Fusarium oxysporum f. sp. lycopersici divides race 1 isolates into separate virulence groups Phytopathology 89 156–160
K O’Donnell HC Kistler E Cigelnik R Ploetz (1998) ArticleTitleMultiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondria gene genealogies Proc Natl Acad Sci USA 95 2044–2049 Occurrence Handle10.1073/pnas.95.5.2044 Occurrence Handle9482835
TL Peever A Ibañez K Akimitsu LW Timmer (2002) ArticleTitleWorldwide phylogeography of the citrus brown spot pathogen, Alternaria alternata Phytopathology 92 794–802
S Pöggeler (1999) ArticleTitlePhylogenetic relationships between mating-type sequences from homothallic and heterothallic ascomycetes Curr Genet 36 222–231 Occurrence Handle10.1007/s002940050494 Occurrence Handle10541860
JE Puhalla (1985) ArticleTitleClassification of strains of Fusarium oxysporum on the basis of vegetative compatibility Can J Bot 63 179–183
MC Ruiz-Roldán A Di Pietro MD Huertas-González MIG Roncero (1999) ArticleTitleTwo xylanase genes of the vascular wilt pathogen Fusarium oxysporum are differentially expressed during infection of tomato plants Mol Gen Genet 261 530–536 Occurrence Handle10.1007/s004380050997 Occurrence Handle10323234
N Saiou M Nei (1987) ArticleTitleThe neighbor-joining method: a new method for reconstructing phylogenetic tree Mol Biol Evol 4 406–425 Occurrence Handle3447015
MB Sela-Buurlage O Budai-Hadrian Q Pan L Carmel-Gore R Vunsch D Zamir R Fluhr (2001) ArticleTitleGenome-wide dissection of Fusarium resistance in tomato reveals multiple complex loci Mol Genet Genomics 265 1104–1111 Occurrence Handle10.1007/s004380100509 Occurrence Handle11523783
T Takehara (1992) ArticleTitleNitrate-nonutilizing mutants of fungi and their use Plant Protection 46 395–399
JD Thompson TJ Gibson F Plewniak F Jeanmougin DG Higgins (1997) ArticleTitleThe Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools Nucleic Acids Res 25 4876–4882 Occurrence Handle10.1093/nar/25.24.4876 Occurrence Handle9396791
BG Turgeon (1998) ArticleTitleApplication of mating type gene technology to problems in fungal biology Annu Rev Phytopathol 36 115–137 Occurrence Handle10.1146/annurev.phyto.36.1.115 Occurrence Handle15012495
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kawabe, M., Kobayashi, Y., Okada, G. et al. Three evolutionary lineages of tomato wilt pathogen, Fusarium oxysporum f. sp. lycopersici, based on sequences of IGS, MAT1, and pg1, are each composed of isolates of a single mating type and a single or closely related vegetative compatibility group. J Gen Plant Pathol 71, 263–272 (2005). https://doi.org/10.1007/s10327-005-0203-6
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/s10327-005-0203-6