Abstract
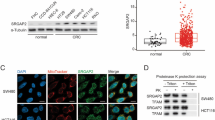
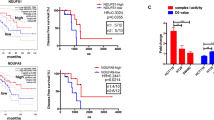
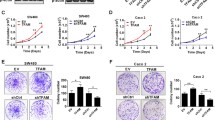
Altered mitochondrial function contributes greatly to pathogenesis and progression of colorectal cancer. In this study, we report a functional pool of Src homology 2 domain-containing F (SHF) in mitochondria controlling the response of colorectal cancer cells to radiation therapy. We found that elevated expression of SHF in cancer cells is essential for promoting mitochondrial function by increasing mitochondrial DNA copy number, thus reducing the sensitivity of colorectal cancer cells to radiation. Mechanistically, SHF binds to mitochondrial DNA and promotes POLG/SSBP1-mediated mitochondrial DNA synthesis. Importantly, SHF loss-mediated radiosensitization was phenocopied by depletion of mitochondrial DNA. Thus, our data demonstrate that mitochondrial SHF is an important regulator of radioresistance in colorectal cancer cells, identifying SHF as a promising therapeutic target to enhance radiotherapy efficacy in colorectal cancer.





Similar content being viewed by others
Data availability
CRC transcriptome sequencing data and corresponding clinical data set were downloaded from TCGA (https://cancergenome.nih.gov, TCGA-COAD and TCGA-READ).
Abbreviations
- CRC:
-
Colorectal cancer
- TCGA:
-
The Cancer Genome Atlas
- OCR:
-
Oxygen consumption rate
- ECAR:
-
Extracellular acidification rate
- MTS:
-
Mitochondrial targeting signal
- NLS:
-
Nuclear localization sequence
- SRB:
-
Sulforhodamine B
- 3’ UTR:
-
3′ Untranslated region
- EV:
-
Empty vector
- ddC:
-
2′,3′-Dideoxycytidine
- PDO:
-
Patient-derived organoid
References
Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72:7–33. https://doi.org/10.3322/caac.21708.
Raza A, et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J Exp Clin Cancer Res CR. 2022;41:99. https://doi.org/10.1186/s13046-022-02318-0.
Brandi G, et al. Is post-transplant chemotherapy feasible in liver transplantation for colorectal cancer liver metastases? Cancer Commun. 2020;40:461–4. https://doi.org/10.1002/cac2.12072.
Rizzo A, et al. Dose reduction and discontinuation of standard-dose regorafenib associated with adverse drug events in cancer patients: a systematic review and meta-analysis. Ther Adv Med Oncol. 2020;12:1758835920936932. https://doi.org/10.1177/1758835920936932.
Huerta S, Gao X, Saha D. Mechanisms of resistance to ionizing radiation in rectal cancer. Expert Rev Mol Diagn. 2009;9:469–80. https://doi.org/10.1586/erm.09.26.
Cui C, et al. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. 2020;19:58. https://doi.org/10.1186/s12943-020-01180-y.
Willems PH, Rossignol R, Dieteren CE, Murphy MP, Koopman WJ. Redox homeostasis and mitochondrial dynamics. Cell Metab. 2015;22:207–18. https://doi.org/10.1016/j.cmet.2015.06.006.
Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98. https://doi.org/10.1038/nrc3365.
Vander-Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–33. https://doi.org/10.1126/science.1160809.
Chekulayev V, et al. Metabolic remodeling in human colorectal cancer and surrounding tissues: alterations in regulation of mitochondrial respiration and metabolic fluxes. Biochem Biophys Rep. 2015;4:111–25. https://doi.org/10.1016/j.bbrep.2015.08.020.
Kaldma A, et al. An in situ study of bioenergetic properties of human colorectal cancer: the regulation of mitochondrial respiration and distribution of flux control among the components of ATP synthasome. Int J Biochem Cell Biol. 2014;55:171–86. https://doi.org/10.1016/j.biocel.2014.09.004.
Sun X, et al. Increased mtDNA copy number promotes cancer progression by enhancing mitochondrial oxidative phosphorylation in microsatellite-stable colorectal cancer. Signal Transduct Target Ther. 2018;3:8. https://doi.org/10.1038/s41392-018-0011-z.
Boyle KA, et al. Mitochondria-targeted drugs stimulate mitophagy and abrogate colon cancer cell proliferation. J Biol Chem. 2018;293:14891–904. https://doi.org/10.1074/jbc.RA117.001469.
Lin CS, et al. Role of mitochondrial function in the invasiveness of human colon cancer cells. Oncol Rep. 2018;39:316–30. https://doi.org/10.3892/or.2017.6087.
Wen YA, et al. The mitochondrial retrograde signaling regulates Wnt signaling to promote tumorigenesis in colon cancer. Cell Death Differ. 2019;26:1955–69. https://doi.org/10.1038/s41418-018-0265-6.
Averbeck D, Rodriguez-Lafrasse C. Role of mitochondria in radiation responses: epigenetic, metabolic, and signaling impacts. Int J Mol Sci. 2021;22:11047. https://doi.org/10.3390/ijms222011047.
Grasso D, et al. Fitter mitochondria are associated with radioresistance in human head and neck SQD9 cancer cells. Front Pharmacol. 2020;11:263. https://doi.org/10.3389/fphar.2020.00263.
Nile DL, et al. Inhibition of glycolysis and mitochondrial respiration promotes radiosensitisation of neuroblastoma and glioma cells. Cancer Metab. 2021;9:24. https://doi.org/10.1186/s40170-021-00258-5.
Qin LL, et al. CDK1 enhances mitochondrial bioenergetics for radiation-induced DNA repair. Cell Rep. 2015;13:2056–63. https://doi.org/10.1016/j.celrep.2015.11.015.
Wang Y, et al. Downregulation of mitochondrial single stranded DNA binding protein (SSBP1) Induces mitochondrial dysfunction and increases the radiosensitivity in non-small cell lung cancer cells. J Cancer. 2017;8:1400–9. https://doi.org/10.7150/jca.18170.
McCann E, O’Sullivan J, Marcone S. Targeting cancer-cell mitochondria and metabolism to improve radiotherapy response. Transl Oncol. 2021;14:100905. https://doi.org/10.1016/j.tranon.2020.100905.
Barchiesi A, Vascotto C. Transcription, processing, and decay of mitochondrial RNA in health and disease. Int J Mol Sci. 2019;20:2221. https://doi.org/10.3390/ijms20092221.
Barshad G, Marom S, Cohen T, Mishmar D. Mitochondrial DNA transcription and its regulation: an evolutionary perspective. Trends Genet. 2018;34:682–92. https://doi.org/10.1016/j.tig.2018.05.009.
Clay-Montier LL, Deng JJ, Bai Y. Number matters: control of mammalian mitochondrial DNA copy number. J Genet Genomic. 2009;36:125–31. https://doi.org/10.1016/S1673-8527(08)60099-5.
Falkenberg M. Mitochondrial DNA replication in mammalian cells: overview of the pathway. Essays Biochem. 2018;62:287–96. https://doi.org/10.1042/Ebc20170100.
Falkenberg M, Gustafsson CM. Mammalian mitochondrial DNA replication and mechanisms of deletion formation. Crit Rev Biochem Mol. 2020;55:509–24. https://doi.org/10.1080/10409238.2020.1818684.
Fontana GA, Gahlon HL. Mechanisms of replication and repair in mitochondrial DNA deletion formation. Nucleic Acid Res. 2020;48:11244–58. https://doi.org/10.1093/nar/gkaa804.
Jiang M, et al. The mitochondrial single-stranded DNA binding protein is essential for initiation of mtDNA replication. Sci Adv. 2021. https://doi.org/10.1126/sciadv.abf8631.
Filograna R, Mennuni M, Alsina D, Larsson NG. Mitochondrial DNA copy number in human disease: the more the better? FEBS Lett. 2021;595:976–1002. https://doi.org/10.1002/1873-3468.14021.
Castellani CA, Longchamps RJ, Sun J, Guallar E, Arking DE. Thinking outside the nucleus: mitochondrial DNA copy number in health and disease. Mitochondrion. 2020;53:214–23. https://doi.org/10.1016/j.mito.2020.06.004.
Jia Y, et al. Lamin B1 loss promotes lung cancer development and metastasis by epigenetic derepression of RET. J Exp Med. 2019;216:1377–95. https://doi.org/10.1084/jem.20181394.
Fujii M, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18:827–38. https://doi.org/10.1016/j.stem.2016.04.003.
Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72. https://doi.org/10.1053/j.gastro.2011.07.050.
Fujii M, Matano M, Nanki K, Sato T. Efficient genetic engineering of human intestinal organoids using electroporation. Nat Protoc. 2015;10:1474–85. https://doi.org/10.1038/nprot.2015.088.
Filograna R, et al. Modulation of mtDNA copy number ameliorates the pathological consequences of a heteroplasmic mtDNA mutation in the mouse. Sci Adv. 2019. https://doi.org/10.1126/sciadv.aav9824.
Venegas V, Halberg MC. Measurement of mitochondrial DNA copy number. Method Mol Biol. 2012;837:327–35. https://doi.org/10.1007/978-1-61779-504-6_22.
Malayil L, et al. Metabolically-active bacteria in reclaimed water and ponds revealed using bromodeoxyuridine DNA labeling coupled with 16S rRNA and shotgun sequencing. Water Res. 2020;184:116185. https://doi.org/10.1016/j.watres.2020.116185.
Malayil L, Chattopadhyay S, Mongodin EF, Sapkota AR. Coupled DNA-labeling and sequencing approach enables the detection of viable-but-non-culturable Vibrio spp. in irrigation water sources in the Chesapeake Bay watershed. Environ Microbiome. 2021;16:13. https://doi.org/10.1186/s40793-021-00382-1.
Cao K, et al. Hypermethylation of hepatic mitochondrial ND6 provokes systemic insulin resistance. Adv Sci. 2021;8:2004507. https://doi.org/10.1002/advs.202004507.
Chatterjee A, et al. MOF acetyl transferase regulates transcription and respiration in mitochondria. Cell. 2016;167:722. https://doi.org/10.1016/j.cell.2016.09.052.
Hansen KG, Herrmann JM. Transport of proteins into mitochondria. Protein J. 2019;38:330–42. https://doi.org/10.1007/s10930-019-09819-6.
Pfanner N, Warscheid B, Wiedemann N. Mitochondrial proteins: from biogenesis to functional networks. Nat Rev Mol Cell Biol. 2019;20:267–84. https://doi.org/10.1038/s41580-018-0092-0.
Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996;241:779–86. https://doi.org/10.1111/j.1432-1033.1996.00779.x.
Fukasawa Y, et al. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomic. 2015;14:1113–26. https://doi.org/10.1074/mcp.M114.043083.
Lee J, et al. Mitochondrial cyclic AMP response element-binding protein (CREB) mediates mitochondrial gene expression and neuronal survival. J Biol Chem. 2005;280:40398–401. https://doi.org/10.1074/jbc.C500140200.
Li M, et al. Identification and characterization of mitochondrial targeting sequence of human apurinic/apyrimidinic endonuclease 1. J Biol Chem. 2010;285:14871–81. https://doi.org/10.1074/jbc.M109.069591.
Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–12. https://doi.org/10.1074/jbc.275.21.16202.
Wiedemann N, Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem. 2017;86:685–714. https://doi.org/10.1146/annurev-biochem-060815-014352.
Chiu HY, et al. Nanoparticle mediated delivery and small molecule triggered activation of proteins in the nucleus. Nucleus. 2018;9:530–42. https://doi.org/10.1080/19491034.2018.1523665.
Dang CV, Lee WM. Identification of the human c-myc protein nuclear translocation signal. Mol Cell Biol. 1988;8:4048–54. https://doi.org/10.1128/mcb.8.10.4048-4054.1988.
Gray M, et al. Development and characterisation of acquired radioresistant breast cancer cell lines. Radiat Oncol. 2019;14:64. https://doi.org/10.1186/s13014-019-1268-2.
Kopinski PK, Singh LN, Zhang S, Lott MT, Wallace DC. Mitochondrial DNA variation and cancer. Nat Rev Cancer. 2021;21:431–45. https://doi.org/10.1038/s41568-021-00358-w.
Merrick BA, et al. HSP binding and mitochondrial localization of p53 protein in human HT1080 and mouse C3H10T1/2 cell lines. Biochem Biophys Acta. 1996;1297:57–68. https://doi.org/10.1016/0167-4838(96)00089-1.
Acknowledgements
We thank the TCGA Research Network for providing its platforms and valuable datasets.
Funding
This present study was funded by a grant from the Young Scientists Fund of the National Natural Science Foundation of China (Grant No. 82203425) and a grant from Shandong Province major science and technology innovation project (Grant no. 2019JZZY011008).
Author information
Authors and Affiliations
Contributions
ZZ, GM, GH, CJ, GJ, and JY were responsible for the concept and design of the original study. ZZ, GM, GH, GJ, and JY analyzed data and illustrated the figures for the manuscript. ZZ, GM, GW, WB, LC, and HQ performed the experiment. ZZ, GM, CJ, GJ, and JY wrote the paper with input from all authors. All authors contributed to the article and approved the submitted version.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Ethical approval and consent to participate
The tissue samples from patients used for the generation of organoids were approved by the ethical review committee of Shandong Cancer Hospital and Institute.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhenyu Zhu and Meihua Gong have contributed equally to this work and share first authorship.
Supplementary Information
Below is the link to the electronic supplementary material.







Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhu, Z., Gong, M., Gong, W. et al. SHF confers radioresistance in colorectal cancer by the regulation of mitochondrial DNA copy number. Clin Exp Med 23, 2457–2471 (2023). https://doi.org/10.1007/s10238-022-00969-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-022-00969-z