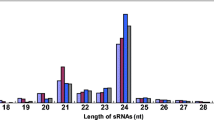
Abstract
Small RNA sequencing (sRNA-seq) and degradome analysis were used for the identification of miRNAs and their target host genes in a pair of near-isogenic lines (NILs), which differed for the presence of leaf rust resistance gene Lr28. The study led to identification of (i) 506 known and 346 novel miRNAs; and (ii) 5054 target genes including 4557 in silico predicted and 497 degradome-based genes using 105 differentially expressed (DE) miRNAs. A subset of 128 targets (67 in silico + 61 degradome-based) was differentially expressed in RNA-seq data that was generated by us earlier using the same pair of NILs; among these 128 targets, 58 target genes exhibited an inverse relationship with the DE miRNAs (expression of miRNAs and activation/suppression of target genes). Eight miRNAs which belonged to the conserved miRNA families and were known to be induced in response to fungal diseases in plants included the following: miR156, miR158, miR159, miR168, miR169, miR172, miR319, miR396. The target genes belonged to the following classes of genes known to be involved in downstream disease resistance pathways; peroxidases, sugar transporters, auxin response signaling, oxidation–reduction, etc. It was also noticed that although a majority of miRNAs and target genes followed the above classical inverse relationship, there were also examples, where no such relationship was observed. Among the target genes, there were also 51 genes that were not only regulated by miRNAs, but were also differentially methylated at sequences including the following segments: promotors, introns, TSS, exons. The results of the present study suggest a complex interplay among miRNA genes, target genes, and various epigenetic controls, which regulate the expression of genes involved in downstream pathways for disease resistance.








Similar content being viewed by others
Data availability
The raw sequence reads data that support the findings of this study are available as Sequence Read Archive of NCBI under accession number PRJNA881058.
References
Afzal F, Chaudhari SK, Gul A, Farooq A, Ali H, Nisar S, Sarfraz B, Shehzadi KJ, Mujeeb-Kazi A (2015) Bread wheat (Triticum aestivum L.) under biotic and abiotic stresses: In: Hakeem K (eds) Crop Prod Global Environ Issues. Springer Cham, pp 293–317. https://doi.org/10.1007/978-3-319-23162-4_13
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Nat Prec 11:R106. https://doi.org/10.1038/npre.2010.4282.1
Balmer A, De Paoli E, Si‐Ammour A, Mauch‐Mani B, Balmer D (2017) Signs of silence: small RNAs and antifungal responses in Arabidopsis thaliana and Zea mays In: Snježana Jurić (ed) Plant Engineering. https://doi.org/10.5772/intechopen.69795
Bao H, Kommadath A, Liang G, Sun X, Arantes AS, Tuggle CK, Bearson S, Plastow GS, Stothard P, Guan LL (2015) Genome-wide whole blood microRNAome and transcriptome analyses reveal miRNA-mRNA regulated host response to foodborne pathogen Salmonella infection in swine. Sci Rep 5(1):1–12. https://doi.org/10.1038/srep12620
Bhavani S, Singh RP, Hodson DP, Huerta-Espino J, Randhawa MS (2022) Wheat rusts: current status, prospects of genetic control and integrated approaches to enhance resistance durability. In: Reynolds MP, Braun HJ (eds) Wheat improvement: food security in a changing climate. Springer Nature Switzerland A, Gewerbestrasse 11, 6330 Cham, Switzerland, 125–141
Bolton MD, Kolmer JA, Garvin DF (2008) Wheat leaf rust caused by Puccinia triticina. Mol Plant Pathol 9(5):563–575. https://doi.org/10.1111/J.1364-3703.2008.00487
Cai Q, Qiao L, Wang M, He B, Lin FM, Palmquist J, Huang SD, Jin H (2018) Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Sci 360:1126–1129. https://doi.org/10.1126/science.aar4142
Chandran V, Wang H, Gao F, Cao XL, Chen YP, Li GB, Zhu Y, Yang XM, Zhang LL, Zhao ZX, Zhao JH, Wang YG, Li S, Fan J, Li Y, Zhao JQ, Li SQ, Wang WM (2019) miR396-OsGRFs module balances growth and rice blast disease resistance. Front Plant Sci 9:1999. https://doi.org/10.3389/fpls.2018.01999
Chen X (2009) Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol 25:21–44. https://doi.org/10.1146/annurev.cellbio.042308.113417
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 619832:1–12. https://doi.org/10.1155/2008/619832
Dai X, Zhuang Z, Zhao PX (2018) psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res 46:49–54. https://doi.org/10.1093/nar/gky316.W49-W54
Deng Y, Ning Y, Yang DL, Zhai K, Wang GL, He Z (2020) Molecular basis of disease resistance and perspectives on breeding strategies for resistance improvement in crops. Mol Plant 13(10):1402–1419. https://doi.org/10.1016/j.molp.2020.09.018
Dhariwal R, Vyas S, Govindraj RB, Jha SK, Khurana JP, Tyagi AK, Prabhu KV, Balyan HS, Gupta PK (2011) Analysis of differentially expressed genes in leaf rust infected bread wheat involving seedling resistance gene Lr28. Functional Plant Biol 38(6):479–492. https://doi.org/10.1071/FP10246
Dhariwal R, Gahlaut V, Govindraj RB, Singh D, Mathur S, Vyas S, Bandopadhyay R, Khurana JP, Tyagi AK, Prabhu KV, Mukhopadhyay K, Balyan HS, Gupta PK (2015) Stage-specific reprogramming of gene expression characterizes Lr48-mediated adult plant leaf rust resistance in wheat. Funct Integr Genom 15:233–245. https://doi.org/10.1007/s10142-014-0416-x
Feng H, Zhang Q, Li H, Wang X, Wang X, Duan X, Wang B, Kang Z (2013) vsiRNAs derived from the miRNA-generating sites of pri-tae-miR159a based on the BSMV system play positive roles in the wheat response to Puccinia striiformis f. sp. tritici through the regulation of taMyb3 expression. Plant Physiol Biochem 68:90–95. https://doi.org/10.1016/j.plaphy.2013.04.008
Feng H, Duan X, Zhang Q, Li X, Wang B, Huang L, Wang X, Kang Z (2014) The target gene of tae-miR164, a novel NAC transcription factor from the NAM subfamily, negatively regulates resistance of wheat to stripe rust. Mol Plant Pathol 15:284–296. https://doi.org/10.1111/mpp.12089
Foster AJ, Jenkinson JM, Talbot NJ (2003) Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J22(2):225–235. https://doi.org/10.1093/emboj/cdg018
Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A (2005) Rfam: annotating non-coding RNAs in complete genomes Nucleic Acid Res 1;33(Database issue):D121–4. https://doi.org/10.1093/nar/gki081
Gupta OP, Permar V, Koundal V, Singh UD, Praveen S (2012) MicroRNA regulated defense responses in Triticum aestivum L. during Puccinia graminis f. sp. tritici infection. Mol Biol Rep 39:817–824. https://doi.org/10.1007/s11033-011-0803-5
Gupta PK (2014) Competing endogenous RNA (ceRNA): a new class of RNA working as miRNA sponges. Curr Sci 106:823–830. https://www.jstor.org/stable/24102268
Gupta PK (2015) miRNAs and target mimics for crop improvement. Curr Sci. 108: 1624–1633. https://www.jstor.org/stable/24905528
Hu Z, Tang Z, Zhang Y, Niu L, Yang F, Zhang D, Hu Y (2021) Rice SUT and SWEET transporters. Int J Mol Sci 22(20):11198. https://doi.org/10.3390/ijms222011198
Inal B, Türktas M, Eren H, Ilhan E, Okay S, Atak M, Erayman M, Unver T (2014) Genome-wide fungal stress responsive miRNA expression in wheat. Planta 240:1287–1298. https://doi.org/10.1007/s00425-014-2153-8
International Wheat Genome Sequencing Consortium (IWGSC)(2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Sci 361:eaar7191. https://doi.org/10.1126/science.aar7191
Jain N, Sinha N, Harikrishna, Singh PK, Gautam T, Prasad P, Balyan HS, Gupta PK (2020) A study of miRNAs and lncRNAs during Lr28-mediated resistance against leaf rust in wheat (Triticum aestivum L.) Physiol Mol Plant Pathol 112:101552. https://doi.org/10.1016/j.pmpp.2020.101552
Jain N, Batra R, Saripalli G, Sinha N, Rani S, Sharma JB, Gautam T, Prasad P, Balyan HS, Gupta PK (2021) Methylome changes during Lr48-mediated APR for leaf rust in wheat (Triticum aestivum L.). Physiol Mol Plant Pathol 116:101726. https://doi.org/10.1016/j.pmpp.2021.101726
Jin W, Wu F (2015) Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biol 15:1–4. https://doi.org/10.1186/s12870-014-0410-4
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799. https://doi.org/10.1016/j.molcel.2004.05.027
Kumar R, Masthigowda MH, Kaur A, Bhusal N, Pandey A, Kumar S, Mishra CN, Singh G, Singh GP (2020) Identification and characterization of multiple abiotic stress tolerance genes in wheat. Mol Biol Rep 47:8629–8643. https://doi.org/10.1007/s11033-020-05906-5
Kumar K, Jan I, Saripalli G, Sharma PK, Mir RR, Balyan HS, Gupta PK (2022) An update on resistance genes and their use in the development of leaf rust resistant cultivars in wheat. Front Genet 13:816057. https://doi.org/10.3389/fgene.2022.816057
Kumar D, Dutta S, Singh D, Prabhu KV, Kumar M, Mukhopadhyay K (2017) Uncovering leaf rust responsive miRNAs in wheat (Triticum aestivum L.) using high-throughput sequencing and prediction of their targets through degradome analysis. Planta 245:161–182. https://doi.org/10.1007/s00425-016-2600-9
Kunkel BN, Johnsons JMB (2021) Auxin plays multiple roles during plant–pathogen interactions. Cold Spring Harb Perspect Biol 1;13(9):a040022. https://doi.org/10.1101/cshperspect.a040022
Li Y, Zhang Z, Liu F, Vongsangnak W, Jing Q, Shen B (2012) Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res 40:4298–4305. https://doi.org/10.1093/nar/gks043
Li Y, Lu YG, Shi Y, Wu L, Xu YJ, Huang F, Guo XY, Zhang Y, Fan J, Zhao JQ et al (2014) Multiple rice microRNAs are involved in immunity against the blast fungus Magnaporthe oryzae. Plant Physiol 164:1077–1092. https://doi.org/10.1104/pp.113.230052
Li Y, Zhao SL, Li JL, Hu XH, Wang H, Cao XL et al (2017) Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front Plant Sci 8:2. https://doi.org/10.3389/fpls.2017.00002
Li D, Yang J, Yang Y, Liu J, Li H, Li R, Cao C, Shi L, Wu W, He K (2021) A timely review of cross-kingdom regulation of plant-derived microRNAs. Front Genet 12:613197. https://doi.org/10.3389/fgene.2021.613197
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lu S, Sun YH, Amerson H, Chiang VL (2007) MicroRNAs in loblolly pine (Pinus taeda L.) and their association with fusiform rust gall development. Plant J 51:1077–98. https://doi.org/10.1111/j.1365-313X.2007.03208.x
Mapuranga J, Zhang N, Zhang L, Liu W, Chang J, Yang W (2022) Harnessing genetic resistance to rusts in wheat and integrated rust management methods to develop more durable resistant cultivars. Front Plant Sci 13:951095. https://doi.org/10.3389/fpls.2022.951095
Meng F, Liu H, Wang K, Liu L, Wang S, Zhao Y, Yin J, Li Y (2013) Development-associated microRNAs in grains of wheat (Triticum aestivum L.). BMC Plant Biol 13:140. https://doi.org/10.1186/1471-2229-13-140
Miao C, Wang D, He R, Liu S, Zhu JK (2020) Mutations in MIR396e and MIR396f increase grain size and modulate shoot architecture in rice. Plant Biotechnol J 18:491–501. https://doi.org/10.1111/pbi.13214
Mutum RD, Kumar S, Balyan S, Kansal S, Mathur S, Raghuvanshi S (2016) Identification of novel miRNAs from drought tolerant rice variety Nagina 22. Sci Rep 6:30786. https://doi.org/10.1038/srep30786
Passardi F, Cosio C, Penel C, Dunand C (2005) Peroxidases have more functions than a Swiss army knife. Plant Cell Rep 24:255–265. https://doi.org/10.1007/s00299-005-09726
Qiu L, Wang H, Li Y, Wang W, Liu Y, Mu J, Geng M, Guo W, Hu Z, Ma J, Sun Q, Xie C (2020) Fine mapping of the wheat leaf rust resistance gene LrLC10 (Lr13) and validation of its co-segregation markers. Front Plant Sci 11:470. https://doi.org/10.3389/fpls.2020.00470
Salvador-Guirao R, Hsing YI, San Segundo B (2018) The polycistronic miR166k- 166h positively regulates rice immunity via post-transcriptional control of EIN2. Front Plant Sci 9:337. https://doi.org/10.3389/fpls.2018.00337
Saripalli G, Sharma C, Gautam T, Singh K, Jain N, Prasad P, Roy JK, Sharma JB, Sharma PK, Prabhu KV, Balyan HS, Gupta PK (2020a) Complex relationship between DNA methylation and gene expression due to Lr28 in wheat-leaf rust pathosystem. Mol Biol Rep 47:1339–1360. https://doi.org/10.1007/s11033-019-05236-1
Saripalli G, Singh K, Gautam T, Kumar S, Raghuvanshi S, Prasad P, Jain N, Sharma PK, Balyan HS, Gupta PK (2020b) Genome-wide analysis of H3K4me3 and H3K27me3 modifications due to Lr28 for leaf rust resistance in bread wheat (Triticum aestivum). Plant Molr Bio 104(1–2):113–136. https://doi.org/10.1007/s11103-020-01029-4
Sarkar D, Maji RK, Dey S, Sarkar A, Ghosh Z, Kundu P (2017) Integrated miRNA and mRNA expression profiling reveals the response regulators of a susceptible tomato cultivar to early blight disease. DNA Res 24(3):235–250. https://doi.org/10.1093/dnares/dsx003
Šečić E, Kogel M-H, Ladera-Carmona MJ (2021) Biotic stress-associated microRNA families in plants. J Plant Physiol 263:153451. https://doi.org/10.1016/j.jplph.2021.153451
Sharma C, Saripalli G, Kumar S, Gautam T, Kumar A, Rani S et al (2018) A study of transcriptome in leaf rust infected bread wheat involving seedling resistance gene Lr28. Funct Plant Biol 45:1046–1064. https://doi.org/10.1071/FP17326
Sharma C, Kumar S, Saripalli G, Jain N, Raghuvanshi S, Sharma JB, Prabhu KV, Sharma PK, Balyan HS, Gupta PK (2019) H3K4/K9 acetylatatin and Lr28-mediated expression of six leaf rust responsive genes in wheat (Triticum aestivum). Mol Genet Genomics 94:227–241. https://doi.org/10.1007/s00438-018-1500-z
Singh K, Saripalli G, Gautam T, Prasad P, Jain N, Balyan HS, Gupta PK (2022) BS-Seq reveals major role of differential CHH methylation during leaf rust resistance in wheat (Triticum aestivum L.). Mol Genet Genomics 297:731–749. https://doi.org/10.1007/s00438-022-01879-1
Springer NM (2013) Epigenetics and crop improvement. Trends Genet 29(4):241–247. https://doi.org/10.1016/j.tig.2012.10.009
Stackman EC, Stewart DM, Loegering WQ (1962) Identification of physiologic races of Puccinia graminis Var. tritici, United States Department of Agriculture. Agricultural Research Service Columbia, MO, USA
Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V (2012) The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinform 28:2059–2061. https://doi.org/10.1093/bioinformatics/bts311
Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genom 160:1–4. https://doi.org/10.1186/1471-2164-9-160
Thody J, Folkes L, Medina-Calzada Z, Xu P, Dalmay T, Moulton V (2018) PAREsnip2: a tool for high-throughput prediction of small RNA targets from degradome sequencing data using configurable targeting rules. Nucleic Acids Res 46:8730–8739. https://doi.org/10.1093/nar/gky609
Vargas WA, Martin JM, Rech GE, Rivera LP, Benito EP, Diaz-Minguez JM, Thon MR, Sukno SA (2012) Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol 158:1342–1358. https://doi.org/10.1104/pp.111.190397
Wang H, Wang H (2015) The miR156/SPL module, a regulatory hub and versatile toolbox, gears up crops for enhanced agronomic traits. Mol Plant 8:677–688. https://doi.org/10.1016/j.molp.2015.01.008
Wang J, Meng X, Dobrovolskaya OB, Orlov YL, Chen M (2017) Non-coding RNAs and their roles in stress response in plants. Genom Proteom Bioinform 15:301–312. https://doi.org/10.1016/j.gpb.2017.01.007
Wu F, Guo Q, Zhang W, Jin W (2015) Identification and analysis of powdery mildew-responsive miRNAs in wheat. J Phytopathol 163:264–270. https://doi.org/10.1111/jph.12315
Xin M, Wang Y, Yao Y, Xic C, Peng H, Ni Z, Sun Q (2010) Diverse set of microRNAs are responsive to powdery mildew infection and heat stress in wheat (Triticum aestivum L.). BMC Plant Biol 10:123. https://doi.org/10.1186/1471-2229-10-123
Ye J, Zhang Y, Cui H, Liu J, Wu Y, Cheng Y, Xu H, Huang X, Li S, Zhou A, Zhang X, Bolund L, Chen Q, Wang J, Yang H, Fang Y, Shi C (2018) WEGO 2.0: a web tool for analyzing and plotting GO annotations. Nucleic Acids Res 46(W1):W71–W75. https://doi.org/10.1093/nar/gky400
Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant miRNA genes. The Plant J 46(2):243–259. https://doi.org/10.1111/j.1365-313X.2006.02697.x
Zhang W, Gao S, Zhou X et al (2011) Bacteria-responsive microRNAs regulate plant innate immunity by modulating plant hormone networks. Plant Mol Biol 75:93–105. https://doi.org/10.1007/s11103-010-9710-8
Zhang T, Zhao YL, Zhao JH, Wang S, Jin Y, Chen ZQ, Fang YY, Hua CL, Ding SW, Guo HS (2016) Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants 2:16153. https://doi.org/10.1038/nplants.2016.153
Zhang Q, Li Y, Zhang Y, Wu C, Wang S, Hao L, Wang S, Li T (2017) Md-miR156ab and Md-miR395 target WRKY transcription factors to influence apple resistance to leaf spot disease. Front Plant Sci 8:526. https://doi.org/10.3389/fpls.2017.00526
Zhang X, Bao Y, Shan D, Wang Z, Song X, Wang Z, Wang J, He J, Wu L, Zhang Z, Niu D, Jin H, Zhao H (2018) Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol 177(1):352–368. https://doi.org/10.1104/pp.17.01665
Zhang LL, Li Y, Zheng YP, Wang H, Yang X, Chen JF, Zhou SX, Wang LF, Li XP, Ma XC, Zhao JQ, Pu M, Feng H, Fan J, Zhang JW, Huang YY, Wang WM (2020) Expressing a target mimic of miR156fhl-3p enhances rice blast disease resistance without yield penalty by improving SPL14 expression. Front Genet 11:327. https://doi.org/10.3389/fgene.2020.00327
Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62:487–495. https://doi.org/10.1093/jxb/erq295
Zogli P, Libault M (2017) Plant response to biotic stress: is there a common epigenetic response during plant-pathogenic and symbiotic interactions? Plant Sci 263:89–93. https://doi.org/10.1016/j.plantsci.2017.07.008
Acknowledgements
We are grateful to NASF, ICAR for providing financial assistance for this study (grant no. NASF/ABP-6006/2016-17). The first author acknowledges the valuable guidance received from Dr. K V Prabhu, Chairman, PPVFRA, India, during this study. HSB was awarded positions of INSA Senior Scientist and later INSA Honorary Scientist during the period of the study.
Funding
The study was supported by financial assistance from the National Agriculture Science Fund, ICAR under grant no. NASF/ABP-6006/2016–17.
Author information
Authors and Affiliations
Contributions
NJ, HSB, and PKG conceived and designed research, wrote and finalized the manuscript. AS, NS carried out wet lab experiments and data analyses, HK, PKS carried out the rust phenotyping work, PP maintained and provided inoculum for leaf rust pathotype 77–5. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jain, N., Shiv, A., Sinha, N. et al. Leaf rust responsive miRNA and their target genes in wheat. Funct Integr Genomics 23, 14 (2023). https://doi.org/10.1007/s10142-022-00928-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10142-022-00928-7