Abstract
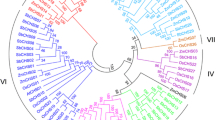
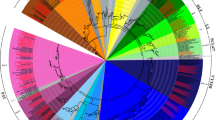
Recent genomic projects reveal that about half of the gene repertoire in plant genomes is made up by multigene families. In this paper, a set of structural and phylogenetic analyses have been applied to compare the differently sized nicotianamine synthase (NAS) gene families in barley and rice. Nicotianamine acts as a chelator of iron and other heavy metals and plays a key role in uptake, phloem transport and cytoplasmic distribution of iron, challenging efforts for the breeding of iron-efficient crop plants. Nine barley NAS genes have been mapped, and co-linearity of flanking genes in barley and rice was determined. The combined analyses reveal that the NAS multigene family members in barley originated through at least one duplication event that occurred before the divergence of rice and barley. Additional duplications appear to have occurred within each of the species. Although we detected no evidence for positive selection of recently duplicated genes within species, codon-based tests revealed evidence for positive selection having contributed to the divergence of some amino acids. The integrated comparative and phylogenetic analysis improved our current view of NAS gene family evolution, might facilitate the functional characterization of individual members and is applicable to other multigene families.






Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. Mol Biol 215:403–410
Bargelloni L, Marcato S, Patarnello T (1998) Antarctic fish hemoglobins: evidence for adaptive evolution at subzero temperature. Proc Natl Acad Sci USA 95:8670–8675
Bauer P, Thiel T, Klatte M, Bereczky Z, Brumbarova T, Hell R, Grosse I (2004) Analysis of sequence, map position, and gene expression reveals conserved essential genes for iron uptake in Arabidopsis and tomato. Plant Physiol 136:4169–4183
Bennett MD, Leitch IJ (1995) Nuclear DNA amounts in angiosperms. Ann Bot 76:113–176
Chen B, Piel HW, Gui L, Bruford E, Monteiro A (2005) The HSP90 family of genes in the human genome: insight into their divergence and evolution. Genomics 86:627–637
Clark RB, Römheld V, Marschner H (1988) Iron uptake and phytosiderophore release by roots of Sorghum genotypes. J Plant Nutr 11(6–11):663–676
Costa JM, Corey A, Hayes PM, Jobet C, Kleinhofs A, Kopisch-Obusch A, Kramer SF, Kudrna D, Li M, Riera-Lizarazu O, Sato K, Szucs P, Toojinda T, Vales MI, Wolfe RI (2001) Molecular mapping of the Oregon Wolfe Barleys: a phenotypically polymorphic doubled-haploid population. Theor Appl Genet 103:415–424
Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409:346–349
Dunford RP, Yano M, Kurata N, Sasaki T, Huestis G, Rocheford T, Laurie DA (2002) Comparative mapping of the barley Ppd-H11 photoperiod response gene region, which lies close to a junction between two rice linkage segments. Genetics 161:825–834
Flavell RB, Bennett MD, Smith JB, Smith DB (1974) Genome size and the proportion of repeated nucleotide sequence DNA in plants. Biochem Genet 12:257–269
Ford MJ (2002) Applications of selective neutrality tests to molecular ecology. Mol Ecol 11:1245–1262
Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95:1971–1974
Gottwald S, Stein N, Boerner A, Sasaki T, Graner A (2004) The gibberellic-acid insensitive dwarfing gene sdw3 of barley is located on chromosome 2HS in a region that shows high colinearity with rice chromosome 7L. Mol Gen Genomics 271:426–436
Graner A, Jahoor A, Schondelmaier J, Siedler H, Pillen K, Fischbeck G, Wenzel G, Herrmann RG (1991) Construction of an RFLP map in barley. Theor Appl Genet 83:250–256
Herbik A, Koch G, Mock HP, Dushkov M, Czihal A, Thielmann J, Stephan UW, Bäumlein H (1999) Isolation, characterization and cDNA cloning of nicotianamine synthase from barley. A key enzyme for iron homeostasis in plants. Eur J Biochem 265:231–239
Higuchi K, Suzuki K, Nakanishi H, Yamaguchi H, Nishizawa NK, Mori S (1999) Cloning of nicotianamine synthase genes, novel genes involved in the biosynthesis of phytosiderophores. Plant Physiol 119:471–479
Higuchi K, Watanabe S, Takahashi M, Kawasaki S, Nakanishi H, Nishizawa NK, Mori S (2001) Nicotianamine synthase gene expression differs in barley and rice under Fe-deficient conditions. Plant J 25:159–167
Islam A, Shepherd KW, Sparrow DHB (1981) Isolation and characterization of euplasmic wheat–barley chromosome addition lines. Heredity 46:161–174
Kawai S, Takagi S, Sato J (1988) Mugineic acid-family phytosiderophores in root-secretions of barley, corn and sorghum varieties. J Plant Nutr 11:633–642
Leister D (2004) Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet 20(3):116–122
Ling HQ, Koch G, Baumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96:7098–7103
Marschner H, Römheld V, Kissel M (1986) Different strategies in higher plants in mobilization and uptake of iron. J Plant Nutr 9:695–713
Mizuno D, Higuchi K, Sakamoto T, Nakanishi, Mori S, Nishizawa NK (2003) Three nicotianamine synthase genes isolated from maize are differentially regulated by iron nutritional status. Plant Physiol 132:1989–1997
Moore G, Devos KM, Wang Z, Gale MD (1995) Cereal genome evolution: grasses, line up and form a circle. Curr Biol 5:737–739
Murata Y, Ma JF, Yamaji N, Ueno D, Nomoto K, Iwashita T (2006) A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J 46:563–572
Palmgren MG (2001) Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annu Rev Plant Physiol Plant Mol Biol 52:817–845
Perovic D, Stein N, Zhang H, Drescher A, Prasad M, Kota R, Kopahnke D, Graner A (2004) An integrated approach for comparative mapping in rice and barley with special reference to the Rph16 resistance locus. Funct Integr Genomics 4:74–83
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Römheld V (1991) The role of phytosiderophores in acquisition of iron and other micronutritients in graminaceous species: an ecological approach. Plant Soil 130:127–134
Römheld V, Marschner H (1986) Evidence for a specific uptake systems for iron phytosiderophores in roots of grasses. Plant Physiol 80:175–180
Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397:694–697
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (Eds.) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 (http://frodo.wi.mit.edu/primer3/primer3.FAQ.html)
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Laboratory Press, New York, pp 2:10–47
Smilde WD, Haluskova J, Sasaki T, Graner A (2001) New evidence for the synteny of rice chromosome 1 and barley chromosome 3H from rice expressed sequence tags. Genome 44:361–367
Sorrells ME, La Rota M, Bermudez-Kandianis CE, Greene RA, Kantety R, Munkvold JD, Miftahudin, Mahmoud A, Ma X, Gustafson PJ, Qi LL, Echalier B, Gill BS, Matthews DE, Lazo GR, Chao S, Anderson OD, Edwards H, Linkiewicz AM, Dubcovsky J, Akhunov ED, Dvorak J, Zhang D, Nguyen HT, Peng J, Lapitan NL, Gonzalez-Hernandez JL, Anderson JA, Hossain K, Kalavacharla V, Kianian SF, Choi DW, Close TJ, Dilbirligi M, Gill KS, Steber C, Walker-Simmons MK, McGuire PE, Qualset CO (2003) Comparative DNA sequence analysis of wheat and rice genomes. Genome Res 13:1818–1827
Stein N, Prasad M, Scholz U, Thiel T, Zhang H, Wolf M, Kota R, Varshney RK, Perovic D, Graner A (2006) A 1000 loci transcript map of the barley genome new anchoring points for integrative grass genomics. Theor Appl Genet (in press)
Stephan UW, Scholz G (1993) Nicotianamine: mediator of iron and heavy metal transport in the phloem? Physiol Plant 88:522–527
Suzuki K, Higuchi K, Nakanashi H, Nishizawa N, Mori S (1999) Cloning of nicotianamine synthase genes from Arabidopsis thaliana. Soil Sci Plant Nutr 45:993–1002
Swoford DL (1998) PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sinauer Associates, Sunderland, MA
Thiel T, Kota R, Grosse I, Stein N, Graner A (2004) SNP2CAPS: a SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res 32:e5
Van Ooijen JW, Voorrips RE (2003) Join Map 3.0 software for the calculation of genetic linkage maps. Plant Research International, Wageningen, The Netherlands
Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14:1223–1233
Von Wirén N, Mori S, Marschner H, Römheld V (1994) Iron inefficiency in maize mutant Ys1 (Zea mays L cv yellow-stripe) is caused by a defect in uptake of iron phytosiderophores. Plant Physiol 106:71–77
World Health Organization (2005) Battling iron deficiency anaemia. http://www.who.int/nut/ida.html
Yang ZH (1998) Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol 15:568–573
Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA (2000) A bacterial artificial chromosome library for barley (Hordeum vulgare L.) and the identification of clones containing putative resistance genes. Theor Appl Genet 101:1093–1099
Zhang H, Sreenivasulu N, Weschke W, Stein N, Rudd S, Radchuk V, Potokina E, Scholz U, Schweizer P, Zierold U, Langridge P, Varshney RK, Wobus U, Graner A (2004) Large-scale analysis of the barley transcriptome based on expressed sequence tags. Plant J 40:276–290
Acknowledgement
We thank Prof. P. Hayes for providing seeds of the OWB mapping population and T. Thiel for support in sequence comparison. P. Tiffin thanks the US National Science Foundation for financial support (DEB 0235027).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the Electronic supplementary material.
Supplemental Figure 1
Multiple alignment of barley NAS genes and positions of designed primers and NASHOR2 oligonucleotide (DOC 51 712 kb)
Supplemental Table 1
Single clone addresses and pools detected after screening of the cv Morex BAC library with NAS genes from barley chromosome 4H. (*) Four common super pools were detected for NASHOR1 and HvNAS2. (**) One common super pool was detected for NASHOR1 and HvNAS3. No common super pools were detected for HvNAS2 and HvNAS3. Single BACs detected by NASHOR1 did not show PCR amplicons with HvNAS2 and HvNAS3 primers (DOC 23 552 kb)
Rights and permissions
About this article
Cite this article
Perovic, D., Tiffin, P., Douchkov, D. et al. An integrated approach for the comparative analysis of a multigene family: The nicotianamine synthase genes of barley. Funct Integr Genomics 7, 169–179 (2007). https://doi.org/10.1007/s10142-006-0040-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-006-0040-5