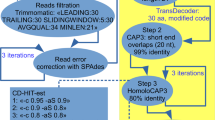
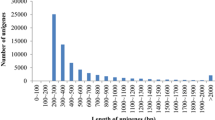
Abstract
Gilthead seabream (Sparus aurata) is a teleost belonging to the family Sparidae with a high economical relevance in the Mediterranean countries. Although genomic tools have been developed in this species in order to investigate its physiology at the molecular level and consequently its culture, genomic information on post-embryonic development is still scarce. In this study, we have investigated the transcriptome of a marine teleost during the larval stage (from hatching to 60 days after hatching) by the use of 454 pyrosequencing technology. We obtained a total of 68,289 assembled contigs, representing putative transcripts, belonging to 54,606 different clusters. Comparison against all S. aurata expressed sequenced tags (ESTs) from the NCBI database revealed that up to 34,722 contigs, belonging to about 61% of gene clusters, are sequences previously not described. Contigs were annotated through an iterative Blast pipeline by comparison against databases such as NCBI RefSeq from Danio rerio, SwissProt or NCBI teleost ESTs. Our results indicate that we have enriched the number of annotated sequences for this species by more than 50% compared with previously existing databases for the gilthead seabream. Gene Ontology analysis of these novel sequences revealed that there is a statistically significant number of transcripts with key roles in larval development, differentiation, morphology, and growth. Finally, all information has been made available online through user-friendly interfaces such as GBrowse and a Blast server with a graphical frontend.



Similar content being viewed by others
References
Adzhubei AA, Vlasova AV, Hagen-Larsen H, Ruden TA, Laerdahl JK, Høyheim B (2007) Annotated Expressed Sequence Tags (ESTs) from pre-smolt Atlantic salmon (Salmo salar) in a searchable data resource. BMC Genomics 8:209
Alami-Durante H, Olive N, Rouel M (2007) Early thermal history significantly affects the seasonal hyperplastic process occurring in the myotomal white muscle of Dicentrarchus labrax juveniles. Cell Tissue Res 327:553–570
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Apromar (2011) La acuicultura marina de peces en España. Fondo Europeo de Pesca, Ministerio de Medio ambiente y Medio Rural y Marino, Spain. 76 pp
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29
Barahona-Fernandez MH (1982) Body deformation in hatchery-reared European seabass Dicentrarchus labrax (L.). Types, prevalence and effect of fish survival. J Fish Biol 21:239–249
Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL (2004) Genomic analysis of mouse retinal development. PLoS Biol 2(9):e247
Bozinovic G, Sit TL, Hinton DE, Oleksiak MF (2011) Gene expression throughout a vertebrate’s embryogenesis. BMC Genomics 12:132
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2008) BLAST+: architecture and applications. BMC Bioinformatics 10:421
Camacho S, Ostos-Garrido MV, Domezain A, Carmona R (2010) Study of the olfactory epithelium in the developing sturgeon. Characterization of the crypt cells. Chem Senses 35:147–156
Campinho MA, Galay-Burgos M, Sweeney GE, Power DM (2010) Coordination of deiodinase and thyroid hormone receptor expression during the larval to juvenile transition in sea bream (Sparus aurata, Linnaeus). Gen Comp Endocrinol 165:181–194
Cerdà J, Mercadé J, Lozano JJ, Manchado M, Tingaud-Sequeira A, Astola A, Infante C, Halm S, Viñas J, Castellana B, Asensio E, Cañavate P, Martínez-Rodríguez G, Piferrer F, Planas J, Prat F, Yúfera M, Durany O, Subirada F, Rosell E, Maes T (2008) Genomic resources for a commercial flatfish, the Senegalese sole (Solea senegalensis): EST sequencing, oligo microarray design, and development of the bioinformatic platform Soleamold. BMC Genomics 9:508
Chevreux B, Wetter T, Suhai S (1999) Genome sequence assembly using trace signals and additional sequence information. Computer Science and Biology. Proceedings of the German Conference on Bioinformatics (GCB) 99:45–56
Chini V, Rimoldi S, Terova G, Saroglia M, Rossi F, Bernardini G, Gornati R (2006) EST-based identification of genes expressed in the liver of adult seabass (Dicentrarchus labrax, L.). Gene 376:102–106
Conesa A, Götz S, García-Gómez J, Terol J, Taron M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinfiormatics 21:3674–3676
Coppe A, Pujolar JM, Maes GE, Larsen PF, Hansen MM, Bernatchez L, Zane L, Bortoluzzi S (2010) Sequencing, de novo annotation and analysis of the first Anguilla anguilla transcriptome: EeelBase opens new perspectives for the study of the critically endangered European eel. BMC Genomics 11:635
Darias MJ, Zambonino-Infante JL, Hugot K, Cahu CL, Mazurais D (2008) Gene expression pattern during the larval development of the European seabass (Dicentrarchus labrax) by microarray analysis. Mar Biotechnol 10:416–428
Douglas SE, Knickle LC, Kimball J, Reith ME (2007) Comprenhensive EST analysis of Atlantic halibut (Hippoglossus hippoglossus), a commercially relevant aquaculture species. BMC Genomics 8:144
Douglas SE, Knickle LC, Williams J, Flight RM, Reith ME (2008) A first generation Atlantic halibut Hippoglossus hippoglossus (L.) microarray: application to developmental studies. J Fish Biol 72:2391–2406
Drivenes Ø, Taranger GL, Edvardsen RB (2012) Gene expression profiling of Atlantic cod (Gadus morhua) embryogenesis using microarray. Mar Biotechnol (in press) doi:10.1007/s10126-011-9399-y
Elmer KR, Fan S, Gunter HM, Jones JC, Boekhoff S, Kuraku S, Meyer A (2010) Rapid evolution and selection inferred from the transcriptomes of sympatric crater lake cichlid fishes. Mol Ecol 19(suppl 1):197–211
Ferrareso S, Vitulo N, Mininni AM, Romualdi C, Cardazzo B, Negrisolo E, Reinhardt R, Canario AVM, Patarnello T, Bargelloni L (2008) Development and validation of a gene expression oligo microarray for the gilthead seabream (Sparus aurata). BMC Genomics 9:580
Ferrareso S, Milan M, Pellizzari C, Vitulo N, Reinhardt R, Canario AVM, Patarnello T, Bargelloni L (2010) Development of an oligo DNA microarray for the European sea bass and its application to expression profiling of jaw deformity. BMC Genomics 11:354
Gibson DA, Ma L (2011) Developmental regulation of axon branching in the vertebrate nervous system. Development 138:183–195
Goetz FW, Rosauer D, Sitar S, Goetz G, Simchick C, Roberts S, Johnson R, Murphy C, Bronte CR, MacKenzie S (2010) A genetic basis for the phenotypic differentiation between siscowet and lean lake trout (Salvelinus namaycush). Mol Ecol 19:176–196
Gonzalez SF, Chatziandreou N, Nielsen ME, Li W, Rogers J, Taylor R, Santos Y, Cossins A (2007) Cutaneous immune responses in the common carp detected using transcript analysis. Mol Immunol 44:1664–1679
Götz S, García-Gómez JM, Terol J, Williams TD, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36(10):3420–3435
Govoroun M, Le Gac F, Guiguen Y (2006) Generation of a large scale repertoire of Expressed Sequence Tags (ESTs) from normalised rainbow trout cDNA libraries. BMC Genomics 7:196
Hagen-Larsen H, Laerdahl JK, Panitz F, Adzhubei A, Høyheim B (2005) An EST-based approach for identifying genes expressed in the intestine and gills of pre-smolt Atlantic salmon (Salmo salar). BMC Genomics 6:171
Hansen A, Zielinski BS (2005) Diversity in the olfactory epithelium of bony fishes: development, lamellar arrangement, sensory neuron cell types and transduction components. J Neurocytol 34:183–208
Hayes B, Laerdahl JK, Lien S, Moen T, Berg P, Hindar K, Davidson WS, Koop BF, Adzhubei A, Høyheim B (2007) An extensive resource of single nucleotide polymorphism markers Associated with Atlantic salmo (Salmo salar) expressed sequences. Aquaculture 265:82–90
Huan P, Wang H, Liu B. (2012) Transcriptomic analysis of the clam Meretrix meretrix on different larval stages. Mar Biotechnol (in press) doi:10.1007/s10126-011-9389-0
Infante C, Asensio E, Cañavate JP, Manchado M (2008) Molecular characterization and expression analysis of five different elongation factors I alpha genes in the flatfish Senegalese sole (Solea senegalensis Kaup): Differential gene expression and thyroid hormones dependence during morphogenesis. BMC Mol Biol 9:19
Jeukens J, Renaut S, St-Cyr J, Nolte AW, Bernatchez L (2010) The transcriptomic of sympatric dwarf and normal lake whitefish (Coregonus clupeaformis spp., Salmonidae) divergence as revealed by next-generation sequencing. Mol Ecol 19:5389–5403
Johansen SD, Karlsen BO, Furmanek T, Andreassen M, Jørgensen TE, Bizuayehu TT, Breines R, Emblem Å, Kettunen P, Luukko K, Edvardsen RB, Nordeide JT, Coucheron DH, Moum T (2011) RNA deep sequencing of Atlantic cod transcriptome. Comp Biochem Physiol part D 6:18–22
Johnston IA (2006) Environment and plasticity of myogenesis in teleost fish. J Exp Biol 209:2249–2264
Johnston IA, Cole NJ, Abercrombie M, Vieira VLA (1998) Embryonic temperature modulates muscle growth characteristics in larval and juvenile herring. J Exp Biol 201:623–646
Koumoundouros G, Ashton C, Sfakianakis DG, Divanach P, Kentouri M, Anthwal N, Stickland NC (2009) Thermally-induced phenotypic plasticity of swimming performance in European sea bass Dicentrarchus labrax juveniles. J Fish Biol 76:1309–1322
Kristiansson E, Asker N, Förlin L, Larsson DGJ (2009) Characterization of the Zoarces viviparous liver transcriptome using massively parallel pyrosequencing. BMC Genomics 10:345
Lamb TD, Collin SP, Pugh EN Jr (2007) Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat Rev Neurosci 8:960–976
Lee BY, Howe AE, Conte MA, D’Cotta H, Pepey E, Baroiller JF, di Palma F, Carleton KL, Kocher TD (2010) An EST resource for tilapia based on 17 normalized libraries and assembly of 116,899 sequence tags. BMC Genomics 11:278
Li P, Peatman E, Wang S, Feng J, He C, Baoprasertkul P, Xu P, Kucuktas H, Nandi S, Somridhivej B, Serapion J, Simmons M, Turan C, Liu L, Muir W, Dunham R, Brady Y, Grizzle J, Liu Z (2007) Towards the ictalurid catfish transcriptome: generation and analysis of 31,215 catfish ESTs. BMC Genomics 8:177
Liu S, Zhou Z, Lu J, Sun F, Wang S, Liu H, Jiang Y, Kucuktas H, Kaltenboeck L, Peatman E, Liu Z (2011) Generation of genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. BMC Genomics 12:53
Louro B, Passos ALS, Souche EL, Tsigenopoulos C, Beck A, Lagnel J, Bonhomme F, Cancela L, Cerdà J, Clark MS, Lubzens E, Magoulas A, Planas JV, Volckaert FAM, Reinhardt R, Canario AVM (2010) Gilthead sea bream (Sparus auratus) and European sea bass (Dicentrarchus labrax) expressed sequence tags: characterization, tissue-specific expression and gene markers. Mar Genomics 3:179–191
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SD, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzik JP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, Mckenna NP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Snirivasan M, Tartaro KR, Tomasz A, Gogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437(7057):376–380
Mazurais D, Darias M, Zambonino-Infante JL, Cahu CL (2011) Transcriptomics for understanding marine fish larval development. Can J Zool 89:599–611
McLean D, Fan J, Higashima SI, Hale M, Fetcho JR (2007) A topographic map of recruitment in spinal cord. Nature 446:71–75
Milan M, Coppe A, Reinhardt R, Cancela LM, Leite RB, Saavedra C, Ciofi C, Chelazzi G, Patarnello T, Bortoluzzi S, Bargelloni L (2011) Transcriptome sequence and microarray development for the Manila clam, Rupitapes philippinarum: genomic tools for environmental monitoring. BMC Genomics 12:234
Morgulis A, Coulouris G, Raytselis Y, Madden TL, Agarwala R, Schäffer AA (2008) Database indexing for production MegaBLAST searches. Bioinformatics 15:1757–1764
Mu Y, Ding F, Cui P, Ao J, Hu S, Chen X (2010) Transcriptome and expression profiling analysis revealed changes of multiple signaling pathways involved in immunity in the large yellow croaker during Aeromonas hydrophila infection. BMC Genomics 11:506
Murray HM, Lall SP, Rajaselvam R, Boutilier LA, Flight RM, Blanchard B, Colombo S, Mohindra V, Yúfera M, Douglas SE (2010) Effect of early introduction of microencapsulated diet to larval Atlantic halibut, Hippoglossus hippoglossus. L. assessed by microarray analysis. Mar Biotechnol 12:214–229
Nieuwenhuys R (2011) The development and general morphology of telescephalon of acnitopterygian fishes: synopsis, documentation and commentary. Brain Struct Funct 215:141–157
Packham IM, Grey C, Heath PR, Hellewell PG, Ingham PW, Crossman DC, Milo M, Chico TJA (2009) Microarray profiling reveals CXCR4a is downregulated by blood flow in vivo and mediates collateral formation in zebrafish embryos. Physiol Genomics 38:319–327
Pardo BG, Fernández C, Millán A, Bouza C, Vázquez-López A, Vera M, Alvarez-Dios JA, Calaza M, Gómez-Tato A, Vázquez M, Cabaleiro S, Magariños B, Lemos ML, Leiro JM, Martínez P (2008) Expressed sequence tags (ESTs) from immune tissues of turbot (Scophthalmus maximus) challenged by pathogens. BMC Vet Res 4:37
Pavlidis M, Mylonas CC (eds.) (2011) Sparidae. Biology and aquaculture of gilthead seabream and other species, Wiley-Blackwell, Oxford. UK. 390 pp
Polo A, Yúfera M, Pascual E (1991) Effect of temperature on egg and larval development of Sparus aurata L. Aquaculture 92:367–375
Polo A, Yúfera M, Pascual E (1992) Feeding and growth of gilthead seabream (Sparus aurata L.) larvae in relation to the size of the rotifer strain used as food. Aquaculture 103:45–54
Power DM, Einarsdóttir IE, Pittman K, Sweeney GE, Hildahl J, Campinho MA, Silva N, Sæle Ø, Galay-Burgos M, Smáradóttir H, Björnsson BT (2008) The molecular and endocrine basis of flatfish metamorphosis. Rev Fish Sci 16(S1):95–111
Renaut S, Nolte AW, Bernatchez L (2010) Mining transcriptome sequences towards identifying adaptive single nucleotide polymorphisms in lake whitefish species pairs (Coregonus spp. Salmonidae). Mol Ecol 19:115–131
Salem M, Rexroad CE III, Wang J, Thorgaard GH, Yao J (2010) Characterization of the rainbow trout transcriptome using Sanger and 454-pyrosequencing approaches. BMC Genomics 11:564
Sánchez CC, Smith TPL, Wiedman RT, Vallejo RL, Salem M, Yao J, Redroad CE III (2009) Single nucleotide polymorphism discovery in rainbow trout by deep sequencing of a reduce representation library. BMC Genomics 10:559
Sandulescu C, Teow RY, Hale ME, Zhang C (2011) Onset and dynamic expression of S100 proteins in the olfactory organ and the lateral line system in zebrafish development. Brain Res 1383:120–127
Sarropoulou E, Power DM, Magoulas A, Geisler R, Kotoulas G (2005a) Comparative analysis and characterization of expressed sequence tags in gilthead sea bream (Sparus aurata) liver and embryos. Aquaculture 243:69–81
Sarropoulou E, Kotoulas G, Power DM, Geisler R (2005b) Gene expression profiling of gilthead seabream during early development and detection of stress-related genes by the application of cDNA microarray technology. Physiol Genomics 23:182–191
Sha Z, Wang S, Zhuang Z, Wang Q, Wang Q, Li P, Ding H, Wang N, Liu Z, Chen S (2010) Generation and analysis of 10 000 ESRs from the half-smooth tongue sole Cynoglossus semilaevis and identification of microsatellite and SNP markers. J Fish Biol 76:1190–1204
Shagin DA, Rebrikov DV, Kozhemyako VB, Altshuler IM, Shcheglov AS, Zhulidov PA, Bogdanova EA, Staroverov DB, Rasskazov VA, Lukyanov S (2002) A novel method for SNP detection using a new duplex-specific nuclease from crab hepatopancreas. Genome Res 12(12):1935–1942
Stein LD, Mungall C, Shu SQ, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, Lewis S (2002) The generic genome browser: a building block for a model organism system database. Genome Res 12:1599–1610
Ton C, Stamatiou D, Dzau VJ, Liew CC (2002) Construction of a zebrafish cDNA microarray: gene expression profiling of the zebrafish during development. Biochem Biophys Res Commun 296:1134–1142
Villeneuve L, Gisbert E, Zambonino-Infante JL, Quazuguel P, Cahu CL (2005) Effect of nature of dietary lipids on European sea bass morphogenesis: implication of retinoid receptors. British J Nutr 94:877–884
Wang S, Peatman E, Abernathy J, Waldbieser G, Lindquist E, Richardson P, Lucas S, Wang M, Li P, Thimmapuram J, Liu L, Vullaganti D, Kucuktas H, Murdock C, Small BC, Wilson M, Liu H, Jiang Y, Lee Y, Chen F, Lu J, Wang W, Xu P, Somridhivej B, Baoprasertkul P, Quilang J, Sha Z, Bao B, Wang Y, Wang Q, Takano T, Nandi S, Liu S, Wong L, Kaltenboeck L, Quiniou S, Bengten E, Miller N, Trant J, Rokhsar D, Liu Z (2010) Assembly of 500,000 inter-specific catfish expressed sequence tags and large scale gene-associated marker development for whole genome association studies. Genome Biol 11:R8
Xia JH, Yue GH (2010) Identification and analysis of immune-related transcriptome in Asian seabass Lates calcarifer. BMC Genomics 11:356
Xiang LX, He D, Dong WR, Zhang YW, Shao JZ (2010) Deep sequencing-based transcriptome profiling analysis of bacteria-challenged Lateolabrax japonicus reveals insight into the immune-relevant genes in marine fish. BMC Genomics 11:472
Yúfera M (2011) Feeding behaviour in larval fish. In: Holt GJ (ed) Larval fish nutrition. Wiley-Blackwell, Ames, pp 285–305
Yúfera M, Darias MJ (2007) The onset of feeding in marine fish larvae. Aquaculture 268:53–63
Yúfera M, Pascual E, Polo A, Sarasquete MC (1993) Effect of starvation on the feeding ability of the gilthead seabream (Sparus aurata L.) larvae at first feeding. J Exp Mar Biol Ecol 169:252–272
Yúfera M, Conceição LEC, Battaglene S, Fushimi H, Kotani T (2011) Early Development and Metabolism. In: Pavlidis M, Mylonas CC (eds) Sparidae. Biology and aquaculture of gilthead seabream and other species. Wiley-Blackwell, Oxford, UK, pp 133–168
Zapata A, Diez B, Cejalvo T, Gutiérrez-de Frías T, Cortés A (2006) Ontogeny of the immune system of fish. Fish Shellfish Immunol 20:126–136
Zhulidov PA, Bogdanova EA, Shcheglov AS, Vagner LL, Khaspekov GL, Kozhemyako VB, Matz MV, Meleshkevitch E, Moroz LL, Lukyanov SA, Shagin DA (2004) Simple cDNA normalization using Kamchatka crab duplex-specific nuclease. Nucleic Acids Res 32(3):e37
Acknowledgments
This research has been funded by the Spanish Ministry of Science and Innovation MICINN + FEDER/ERDF, Consolider Ingenio 2010 Program (Project Aquagenomics, CSD2007-0002). This study also benefited from participation in LARVANET-COST action FA0801 (EU RTD framework Programme). We would like to thank V. Gordo, P. Altube, C. Ligero and A. Godoy for their help in implementing GBrowse and Blast servers.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yúfera, M., Halm, S., Beltran, S. et al. Transcriptomic Characterization of the Larval Stage in Gilthead Seabream (Sparus aurata) by 454 Pyrosequencing. Mar Biotechnol 14, 423–435 (2012). https://doi.org/10.1007/s10126-011-9422-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10126-011-9422-3