Abstract
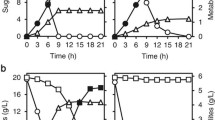
Genome shuffling by recursive protoplast fusion between Saccharomyces cerevisiae and Pichia stipitis also known as Scheffersomyces stipitis resulted in a promising yeast hybrid strain with superior qualities than those of the parental strains in enhancing biofuel production. Our study focused on the substrate utilization, ethanol fermentation, and ethanol tolerance of the hybrids and the parental strains. The parental strain S. cerevisiae is limited to utilize only hexose sugars, and this leads to decrease in the ethanol yield when they are subjected to ethanol production from lignocellulosic biomass which is rich in pentose sugars. To overcome this limitation, we constructed a hybrid yeast strain through genome shuffling which can assimilate all the sugars present in the fermentation medium. After two rounds of recursive protoplast fusion, there was a higher increase in substrate utilization by hybrid SP2-18 compared to parental strain S. cerevisiae. SP2-18 was able to consume 34% of xylose sugar present in the fermentation medium, whereas S. cerevisiae was not able to utilize xylose. Further, the hybrid strain SP2-18 was able to reach an ethanol productivity of 1.03 g L−1 h−1, ethanol yield 0.447 g/g, and ethanol concentration 74.65 g L−1 which was relatively higher than that of the parental strain S. cerevisiae. Furthermore, the hybrid SP2-18 was found to be stable in the production of ethanol. The random amplified polymorphic DNA profile of the yeast hybrid SP2-18 shows the polymorphism between the parental strains indicating the migration of specific sugar metabolizing genes from P. stipitis, while the maximum similarity was with the parent S. cerevisiae.





Similar content being viewed by others
References
Aro EM (2016) From first generation biofuels to advanced solar biofuels. Ambio 45:24–31
Chen L, Zhang J, Chen WN (2014) Engineering the Saccharomyces cerevisiae β-oxidation pathway to increase medium chain fatty acid production as potential biofuel. PLoS One 9(1):e84853. https://doi.org/10.1371/journal.pone.0084853
Coyle W (2007) The future of biofuels a global perspective. Europe 5:24–29
Curran BP, Bugeja VC (1996) Protoplast fusion in Saccharomyces cerevisiae. Methods Mol Biol:45–49
del Cardayré SB (2005) Developments in strain improvement technology. Nat Prod:117–119
El-Fiky ZA, Hassan GM, Emam AM (2012) Quality parameters and RAPD-PCR differentiation of commercial Baker’s yeast and hybrid strains. J Food Sci 77:M312–M317. https://doi.org/10.1111/j.1750-3841.2012.02690.x
Elsh J, Mcclelland M (1991) Genomic fingerprinting using arbitrarily primed PCR and a matrix of pairwise combinations of primers. Nucleic Acids Res 19:5275–5279
Gong J, Zheng H, Wu Z, Chen T, Zhao X (2009) Genome shuffling: progress and applications for phenotype improvement. Biotechnol Adv 27:996–1005
Harju S, Fedosyuk H, Peterson KR (2004) Rapid isolation of yeast genomic DNA: Bust n’ Grab. BMC Biotechnol 4(8)
Jetti K, Nammi S (2017) Construction of xylose assimilating yeast hybrids through genome shuffling. Int J Pharma Bio Sci 8:873–881
Jingping G, Hongbing S, Gang S, Hongzhi L, Wenxiang P (2012) A genome shuffling-generated Saccharomyces cerevisiae isolate that ferments xylose and glucose to produce high levels of ethanol. J Ind Microbiol Biotechnol 39(5):777–787
John RP, Gangadharan D, Madhavan Nampoothiri K (2008) Genome shuffling of Lactobacillus delbrueckii mutant and Bacillus amyloliquefaciens through protoplasmic fusion for L-lactic acid production from starchy wastes. Bioresour Technol 99:8008–8015
Kavanagh K, Whittaker PA (1996) Application of protoplast fusion to the nonconventional yeast. Enzym Microb Technol 18:45–51
Kiransree N, Sridhar M, Venkateswar Rao L (2000) Characterisation of thermotolerant, ethanol tolerant fermentative Saccharomyces cerevisiae for ethanol production. Bioprocess Eng 22:0243–0246
Lebeau T, Jouenne T, Junter GA (2007) Long-term incomplete xylose fermentation, after glucose exhaustion, with Candida shehatae co-immobilized with Saccharomyces cerevisiae. Microbiol Res 162:211–218
Liu Q, Kane PM, Newman PR, Forgac M (1996) Site-directed mutagenesis of the yeast V-ATPase B subunit (Vma2p). J Biol Chem 271:2018–2022
Matsushika A, Sawayama S (2008) Efficient bioethanol production from xylose by recombinant Saccharomyces cerevisiae requires high activity of xylose reductase and moderate xylulokinase activity. J Biosci Bioeng 106:306–309
Naik SN, Goud VV, Rout PK, Dalai AK (2010) Production of first and second generation biofuels: a comprehensive review. Renew Sust Energ Rev 14:578–597
Otte B, Grunwaldt E, Mahmoud O, Jennewein S (2009) Genome shuffling in Clostridium diolis DSM 15410 for improved 1,3-propanediol production. Appl Environ Microbiol 75:7610–7616
Pang ZW, Liang JJ, Qin XJ, Wang JR, Feng JX, Huang RB (2010) Multiple induced mutagenesis for improvement of ethanol production by Kluyveromyces marxianus. Biotechnol Lett 32:1847–1851
Patnaik R, Louie S, Gavrilovic V, Perry K, Stemmer WPC, Ryan CM, del Cardayré S (2002) Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol 20:707–712
Sánchez ÓJ, Cardona CA (2008) Trends in biotechnological production of fuel ethanol from different feedstocks. Bioresour Technol 99:5270–5295
Stephanopoulos G (2002) Metabolic engineering by genome shuffling - two reports on whole-genome shuffling demonstrate the application of combinatorial methods for phenotypic improvement in bacteria. Nat Biotechnol 20:666–668
Sukumaran RK, Surender VJ, Sindhu R, Binod P, Janu KU, Sajna KV, Rajasree KP, Pandey A (2010) Lignocellulosic ethanol in India: prospects, challenges and feedstock availability. Bioresour Technol 101:4826–4833
Swan TM, Watson K (1999) Stress tolerance in a yeast lipid mutant: membrane lipids influence tolerance to heat and ethanol independently of heat shock proteins and trehalose. Can J Microbiol 45:472–479
Van Maris AJA, Winkler AA, Kuyper M, De Laat WTAM, Van Dijken JP, Pronk JT (2007) Development of efficient xylose fermentation in saccharomyces cerevisiae: xylose isomerase as a key component. Adv Biochem Eng Biotechnol 108:179–204
Wang DSCWK (2009) Genome shuffling to improve thermotolerance, ethanol tolerance and ethanol productivity of Saccharomyces cerevisiae. J Ind Microbiol Biotechnol 36(1):139-147.
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18:6531–6535
Yamaoka C, Kurita O, Kubo T (2014) Improved ethanol tolerance of Saccharomyces cerevisiae in mixed cultures with Kluyveromyces lactis on high-sugar fermentation. Microbiol Res 169:907–914
Yan F, Bai F, Tian S, Zhang J, Zhang Z, Yang X (2009) Strain construction for ethanol production from dilute-acid lignocellulosic hydrolysate. Appl Biochem Biotechnol 157:473–482
Zabed H, Sahu JN, Boyce AN, Faruq G (2016) Fuel ethanol production from lignocellulosic biomass: an overview on feedstocks and technological approaches. Renew Sust Energ Rev 66:751–774
Zhang W, Geng A (2012) Improved ethanol production by a xylose-fermenting recombinant yeast strain constructed through a modified genome shuffling method. Biotechnol Biofuels 5:46
Zhang Y-X, Perry K, V a V, Powell K, Stemmer WPC, del Cardayré SB (2002) Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature 415:644–646
Zhang Y, Liu JZ, Huang JS, Mao ZW (2010) Genome shuffling of Propionibacterium shermanii for improving vitamin B12 production and comparative proteome analysis. J Biotechnol 148:139–143
Acknowledgments
Authors are grateful to GITAM deemed to be University, Visakhapatnam for providing all the facilities.
Funding
This work is supported by a grant awarded by the Department of Science and Technology (DST), under Women Scientist Scheme—A (WOS-A), Order No. SR/WOS-A/LS-1214/2015 (G) dt. .12.09.2016 to the first author.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Rights and permissions
About this article
Cite this article
Jetti, K.D., GNS, R.R., Garlapati, D. et al. Improved ethanol productivity and ethanol tolerance through genome shuffling of Saccharomyces cerevisiae and Pichia stipitis. Int Microbiol 22, 247–254 (2019). https://doi.org/10.1007/s10123-018-00044-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10123-018-00044-2