Abstract
Background
Metastatic gastric cancer (GC) has a poor prognosis, and elucidating the molecular mechanisms involved in metastasis may lead to the development of novel therapeutic modalities.
Methods
Transcriptome analysis of surgically resected metastatic tissue from GC patients and noncancerous tissue was performed to identify novel metastasis-related genes. Analyses of in vitro cell function, apoptosis, the cell cycle and cancer stemness were performed using GC cell lines with a stable knockout of a candidate gene. In vivo percutaneous, peritoneal dissemination and liver metastasis xenograft models were also generated. PCR array and proteome analyses were performed. Expression of the candidate gene was analyzed in GC tissues from 300 patients.
Results
Lysosomal Associated Membrane Protein Family Member 5 (LAMP5) was upregulated in the metastatic tissues. LAMP5 knockout significantly suppressed proliferation, invasion, and migration of GC cells and increased apoptosis, cell cycle arrest and cancer stemness. LAMP5 knockout virtually suppressed tumor growth in in vivo percutaneous, peritoneal dissemination and liver metastasis models. EMT- and autophagy-related genes were associated with LAMP5. High LAMP5 mRNA levels were significantly associated with a worse prognosis.
Conclusion
LAMP5 plays a vital role in metastasis formation and may be a promising novel target of drug development for metastatic GC in the future.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is one of the most common malignant tumours and has become a major cause of cancer-related death worldwide [1]. New treatment strategies are needed for advanced GC because advanced GC patients experience recurrence with a high probability, and the mortality rate also remains high after relapse [2, 3]. The development of predictive markers for tumour recurrence, diagnostic markers of recurrence and therapeutic agents is needed to improve the prognosis of advanced GG patients [4]. As trastuzumab, ramucirumab and nivolumab have been developed as molecularly targeted drugs to inhibit GC progression [5,6,7], the elucidation of the mechanisms and identification of molecules related to GC recurrence will promote the development of new molecular targeted drugs for metastatic GC, including the prevention of metastasis.
The process of metastatic site formation from the primary region includes epithelial-mesenchymal transition (EMT) [8], invasion to the circulation, migration and proliferation in circulation[9], spheroid formation and proliferation in the microenvironment of the target organ [10], and multistep alteration of gene expression from the primary region to the metastatic site may be involved in the expression of these cancer phenotypes[11]. Differences in gene expression between primary and metastatic regions have been reported [12]. Therefore, important molecules related to metastasis of GC may be undetectable with the examination of the primary region only, and analysis of metastatic tissues is needed for further identification of these molecules.
In this study, global messenger RNA expression analysis focusing on metastatic tissue from GC patients was performed, and Lysosomal Associated Membrane Protein Family Member 5 (LAMP5), which is overexpressed only in metastatic sites, was identified. LAMP5 is a single-pass transmembrane protein and has a role in lysosomal function. We established a stable LAMP5 knockout (KO) GC cell line and analyzed various alterations in the malignant phenotype associated with metastasis of GC to reveal the role of LAMP5 in GC progression.
Materials and methods
Global mRNA expression analysis
Global expression profiling was conducted to compare the expression levels of 57,749 genes between primary lesion, liver metastasis and adjacent normal tissues was performed (n = 4) by the HiSeq platform (Illumina, San Diego, CA, USA) with 100 real length and paired-end read. The qualities of the RNA preparations were sufficient for analysis indicated as follows: yield data per sample = 2980 Mb, mean reads per sample = 29,042,165 pairs, mean rate ≥ Q30 = 94.4%, mean quality score = 36.5 and mean total mapped-read rate = 93.1%. The sequencing coverage and quality statistics for each sample are summarized in supplementary table 1. We filtered 57,749 genes to yield 14 genes according to three criteria: (1) no significant difference (P > 0.05) between primary cancer lesions (C) and adjacent normal gastric tissues (N), (2) up regulated in liver metastatic site (H) (3) significant difference (Q < 0.05) between H and N.
Cell lines
MKN1(RRID:CVCL_1415), MKN7 (RRID:CVCL_1417), MKN45 (RRID:CVCL_0434), MKN74 (RRID:CVCL_2791), IM95 (RRID:CVCL_2961), OCUM1 (RRID:CVCL_3084), RERF-GC-1Bb, SC-6-JCK(RRID:CVCL_F953), H-111-TC (RRID:CVCL_5511), HS746T (RRID: CVCL_0333), FU97 (RRID: CVCL_2908), GSU (RRID: CVCL_8877), HGC27 (RRID: CVCL_1279), and SH-10-TC (RRID:CVCL_5167) cell lines were purchased from the collection of the Research Bioresources Cell Bank (JCRB, Osaka, Japan). FHs74 (RRID: CVCL_2899), AGS (RRID: CVCL_0139), KATOIII (RRID: CVCL_M251), and N87 (RRID: CVCL_1603) cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA), and GCIY (RRID: CVCL_1228) was obtained from the Cell Resource Center for Biomedical Research/Cell Bank (Sendai, Japan). NUGC2 (RRID: CVCL_1611), NUGC3 (RRID: CVCL_1612), and NUGC4 (RRID: CVCL_3082) cell lines were established in our institution. All human cell lines have been authenticated using STR profiling within the last three years and all experiments were performed with mycoplasma-free cells.
Determination of mRNA expression levels
mRNA expression levels were evaluated by quantitative reverse transcription-polymerase chain reaction (q-PCR) as previously described[13]. GAPDH was employed as the endogenous control to normalize the expression levels[14]. Primer and probe DNA sequences are shown in Supplementary Table 2.
Establishment of stable KO cell lines
We selected the MKN1 cell lines for LAMP5-KO, because they expressed relatively high levels of LAMP5 and were known to be suitable for a stable knockout, cell function assays, flow cytometric assays and in vivo experiments analysis from our previous studies[15, 16].
KO cell lines were established by CRISPR/Cas9 system. Guide RNA (gRNA) target sequences were determined using the Gene Art CRISPR gRNA Design Tool (Thermo Fisher Scientific, Waltham, MA, USA), and gRNA was synthesized with a Precision gRNA Synthesis Kit (Thermo Fisher Scientific). Synthesized RNA (200 ng), Gene Art Platinum Cas9 nuclease (0.83 µg) (Thermo Fisher Scientific) and 1 × 106 cells were mixed with R-buffer (100 µl) (Thermo Fisher Scientific) and transfected using the NEON electroporation system (1400 V, pulse width 20 ms, two pulses) (Thermo Fisher Scientific). Cleavage detection was performed 48 h after transfection with a Gene Art Genomic Cleavage Detection Kit (Thermo Fisher Scientific), and fragmentation was observed (Fig. 1B).
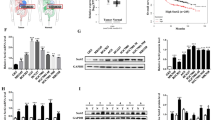
LAMP5 mRNA expression is upregulated in GC cell lines, and LAMP5 knock out (KO) inhibits cell proliferation, controlling cell apoptosis and the cell cycle. a LAMP5 mRNA expression in a noncancerous intestinal cell line (FHS74) and 21 GC cell lines. b Cleavage detection of two kinds of LAMP5-KO cells. c LAMP5 protein expression analysis by western blotting in MKN1 cells and two kinds of LAMP5-KO cells. d Proliferation of MKN1 and 2 kinds of LAMP5-KO cells. The fold changes from day 0 are shown (*P < 0.05). e Analysis of mitochondrial membrane depolarization in MKN1 and two kinds of LAMP5-KO cells. The percentages of depolarized cells and dead cells are shown. f Analysis of the cell cycle of MKN1 and two kinds of LAMP5-KO cells. The percentages of G1-, S-, and G2/M-phase cells are shown in the upper right figures. g Invasion assays of MKN1 and two kinds of LAMP5-KO cells. The graph shows the number of cells that invaded the Matrigel chamber per section
One hundred microliters (5 cells/1 ml) of transfected cells were seeded in a 96-well plate, and the cells that formed a single colony were subcultured to establish a stable KO cell line. The genome-editing site was cloned by a TOPO TA cloning kit (Thermo Fisher Scientific), and sequencing was performed by Eurofins Genomics Co., Ltd., (Tokyo, Japan) using a Big Dye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific) and a 3730 × l DNA Analyzer (Applied Biosystems) [15]. Primer sequences for the cleavage assay and gRNA target sequences were described (Supplementary Table 2).
Small interfering RNA-mediated knockdown of LAMP5
It is important to determine whether the results of experiments with MKN1 are reproducible in other types of cell lines to show the general function of LAMP5 in GC. A specific small interfering RNA (siRNA)-mediated knockdown of LAMP5 expression was performed to evaluate the effect of LAMP5 inhibition on proliferation activity of multiple GC cells; N87, a differentiated gastric adenocarcinoma cell line, and NUGC3 and NUGC4, poorly differentiated gastric adenocarcinoma cell lines. N87, NUGC3 and NUGC4 cells were cultured in a 24-well plate (3 × 104 cells/ml), and cells were transiently transfected the next day with 30 nM siRNA specific for LAMP5 (Supplementary Table 2) or a control siRNA (siControl) combined with LipoTrust EX Oligo (Hokkaido System Science, Sapporo, Japan). After transfection, cells were cultured in serum-free RPMI for 72 h and measured the KD efficacy by q-PCR and used in functional assays.
Cell proliferation assay
Cell proliferation was detected by Cell Counting Kit-8 assays (Dojindo Molecular Technologies, Inc., Kumamoto, Japan). A total of 3 × 103 cells were seeded in 96-well plates and measured on days 0, 1, 3, 5, and 7, and the ratio of change from day 0 was calculated.
Apoptosis, caspase, cell cycle and oxygen stress assays
Total caspase activation was detected by a Muse Multicaspase Kit (Merck Millipore, Billerica, MA, USA), and individual caspase activation was measured by a Caspase-3 Fluorometric Assay Kit, Caspase-8 Fluorometric Assay Kit, Caspase-9 Fluorometric Assay Kit and Caspase-12 Fluorometric Assay Kit (BioVision) using 1 × 106 cells for each assay according to the protocol. Mitochondrial membrane potential was measured by a Muse® Mitopotential Assay Kit (Merck). The cell cycle was determined using a Muse Cell Cycle Kit according to the protocol. Furthermore, oxidative stress was detected by a Muse Oxidative Stress Kit (Merck).
Cell function analysis
Invasion assays were performed with BioCoat Matrigel invasion chambers (BD Biosciences, Bedford, MA, USA). A total of 2.5 × 104 cells were seeded in a Matrigel chamber with FBS-negative medium, and invading cells were stained and counted 48 h after dissemination. A wound-healing assay was conducted with Culture-Insert 2 Well in a µ-Dish (35 mm, ibidi). A total of 3 × 104 cells resuspended in 70 µl of medium were seeded in each column, and after 24 h of incubation, the insert was removed, and the migratory distance was measured every 8 h with an FSX100 (Olympus, Tokyo, Japan) microscope.
Cancer stemness assay
The cancer stemness marker aldehyde dehydrogenase (ALDH) was detected using an ALDEFLUOR™ Kit (StemCell). Then, 500 µl of a 5 × 105 cells/ml cell suspension was added to a test tube and negative control tube, 5 µl of DEAB reagent was added to a negative control tube, and 2.5 µl of ALDEFLUOR™ Reagent was added to each tube. After 45 min of incubation, FL1 (ALDEFLUOR™ Fluorescence) was analysed by flow cytometry using a BD FACS Calibur system (BD Biosciences).
Spheroid formation analysis
Spheroid formation was analyzed using a Cultrex 3D Spheroid Proliferation/Viability Assay Kit (R&D Systems, Inc.) according to the protocol. A total of 3 × 103 cells were incubated with ECM for 72 h to form spheroids. Then, images of spheroids were captured at day 0, day 1, day 3 and day 5 by FSX100 at 40 ×, and the area of the spheroids was measured by ImageJ. The viability of the spheroids was detected by MMT assays.
Pathway analysis
A Human XL Oncology Array Kit from the Proteome Profiler Antibody Arrays (R&D Systems, Minneapolis, MN) was used to determine the relative expression levels of 84 human cancer-related proteins in MKN1 cells and KO cells. mRNA expression analysis of 84 EMT-related cancer pathway molecules was performed by RT2 Profiler PCR Arrays (Qiagen, Hilden, Germany). The molecules that showed correlations in expression levels in 14 cell lines were identified, and changes induced by KO were validated. Autophagy-related molecules were analysed by RT2 profiler PCR arrays using parent cells and KO cells.
Mouse subcutaneous xenograft model
The Animal Research Committee of Nagoya University approved all animal experiments. A total of 1 × 106 cells resuspended in 50 µl of PBS and 50 µl of Matrigel (Corning) were injected into the bilateral backs of 6-w/o male BALBc (nu/nu) mice (n = 4) (SLC, Inc., Hamamatsu, Japan). Tumour size was measured every week, and tumours were collected after sacrifice 8 weeks after implantation. Tumour volume was calculated as length × width × height.
Mouse peritoneal dissemination model
A total of 1 × 106 cells were resuspended in 1 ml of PBS and injected into the peritoneal cavity of 6 w/o male BALBc (nu/nu) mice (n = 5) (SLC, Inc., Hamamatsu, Japan). Tumour growth was followed by an In Vivo Imaging System (IVIS) (Xenogen, Alameda, CA, USA) every 2 weeks after dissemination. Mice were sacrificed 8 weeks after implantation. The total tumour volume was evaluated[17].
Liver metastasis model
Six-week-old male NOD SCID mice (CLEA, Japan) were used (n = 4). A total of 5 × 105 cells resuspended in 100 µl of PBS were injected slowly into the portal vein for 60 s with a 35G injector under laparotomy and a magnifying glass. Tumour formation was confirmed by an In Vivo Imaging System (IVIS) (Xenogen, Alameda, CA, USA) every 4 weeks after tumour injection. D-luciferin (150 mg/kg) was administered intraperitoneally, and photon quantities were measured after 15 min. Living Image Ver. 2.6 software (Xenogen) was used to analyse the IVIS data. Twelve weeks after injection, magnetic resonance imaging (MRS 3000, MR solutions, Guildford, UK) was conducted to observe tumorigenesis, and the tumours were collected after sacrifice as scheduled. Liver specimens were fixed with formalin and sliced in the coronal plane at 2 mm intervals to investigate tumour formation[18].
Analysis of clinical samples
The records of 300 GC patients who underwent surgical resection between 2001 and 2017 were analysed. The LAMP5 mRNA levels in the primary region and adjacent noncancerous tissue were measured using qRT-PCR assays. Correlations between LAMP5 mRNA levels and clinicopathological findings and prognosis were evaluated. Written informed consent was obtained from all patients. The Nagoya University ethics committee approved this study. Clinical staging was performed according to the UICC classification (7th edition). After 2006, patients were administered adjuvant S-1 chemotherapy, if possible.
Immunohistochemistry
Immunohistochemical staining was performed to determine the difference in LAMP5 protein expression between the liver metastatic region and primary region of 17 clinical cases with resection of liver metastasis. The sections were incubated for 16 h at 4 °C with a rabbit polyclonal antibody raised against LAMP5 (HPA031369, Atlas Antibodies, Stockholm, Sweden) diluted 1:100 in antibody diluent (Dako, Carpinteria, CA, USA). The sections were incubated with biotinylated secondary antibody (SignalStain® Boost IHC Detection Reagent labelled by HRP, Cell Signaling Technology, Beverly, MA, USA) for 30 min. Antigen-antibody complexes were visualized by exposure to liquid 3,3′-diaminobenzidine (Nichirei, Tokyo, Japan) for two minutes. Two independent observers graded staining intensity of primary region and metastasis region depending on the percentage of stained cells as follows: 0 (no staining), 1 + (< 10%), 2 + (10 ~ 30%) and 3 + (30% <).
Statistical analysis
The Mann–Whitney test was employed to determine the significance of the difference in the average values of two groups, and the χ2 test was used to compare the ratio of two groups. Spearman's correlation coefficient was employed to test the paired-bivariate correlation. The cumulative recurrence rate was determined by the Kaplan–Meier method, and significant differences between the two groups were tested by the log-rank test. Univariable and multivariable analyses were performed with the Cox hazards ratio. In any tests, P < 0.05 was considered to indicate statistical significance. JMP 13 software (SAS Institute, Inc., Cary, NC, USA) was employed for statistical analyses.
Results
Global mRNA analysis
Twelve candidate genes whose mRNA expression in metastatic tissue was increased compared to that in noncancerous stomach tissue were identified as candidates for metastasis-related genes in GC. Among them, LAMP5 was selected for further study because its role in digestive cancer has not been reported previously (Table 1).
LAMP5 expression levels in GC cell lines
The LAMP5 mRNA levels in 20 GC cell lines varied, and the LAMP5 mRNA levels in 12 cell lines were higher than that in FHS74 (Fig. 1A). There was no relation between LAMP5 expression and cell line differentiation. MKN1 was chosen for LAMP5 KO with CRISPR Cas9.
Confirmation of LAMP5 KO
MKN1 was knocked out using two kinds of guide RNAs, and two kinds of stable LAMP5-KO cell lines were established. Cleavage bands were detected in both kinds of KO cells (Fig. 1B), and a decrease in LAMP5 protein expression was proven by western blotting analysis (Fig. 1C).
Proliferation, apoptosis, cell cycle and ROS activation
The proliferation of KO-LAMP5-1 and KO-LAMP5-2 cells 72 h and 120 h after dissemination was decreased compared to that of MKN1 cells (Fig. 1D). Total caspase activation was increased in the KO-LAMP5-2 cells but not in the KO-LAMP5-1 cells (Supplementary Fig. 1A), and mitochondrial membrane depolarization was detected in both LAMP5 KO cell lines (Fig. 1E). Individual caspase assays revealed that caspase 3 and caspase 9 were increased by LAMP5 KO; however, caspase 8 and caspase 12 were not changed (Supplementary Fig. 1B). The proportion of cells in G1 phase was decreased by LAMP5 KO, and the proportions of cells in S phase and G2/M phase were increased in both KO cell lines (Fig. 1F). Increased ROS activation in the KO-LAMP5-2 cells relative to the parent cells was detected (Supplementary Fig. 1C).
Cell function analysis
Invasion of KO-LAMP5-1 and KO-LAMP5-2 cells was significantly decreased compared to that of the parent cells (Fig. 1G). Migration was also decreased by LAMP5 KO in both cell lines. The suppressive effect on migration induced by LAMP5 was stronger in the KO-LAMP5-1 cells than in the KO-LAMP5-2 cells (Fig. 2A).
LAMP5 knockout influences the migration, cancer stemness and spheroid formation of GC cells. a Images (left) and quantification (right) of the results of wound-healing migration assays of MKN1 and two kinds of LAMP5-KO cells. b Cancer stemness assays of MKN1 and two kinds of LAMP5-KO cells. The percentage of ALDH-positive cells is shown in the lower right of the figures. Negative control (upper) and stemness detection (lower) are shown. c Images of spheroids at day 0, day 1, day 3 and day 5 are shown (left), and the graph shows the mean spheroid area (right)
Cancer stemness and spheroid formation
A decrease in the cancer stemness in the KO-LAMP5-2 cells compared to the parent cells was shown by ALDH assays (Fig. 2B). Spheroid formation was also decreased by LAMP5 KO, and the spheroid shape collapsed in both LAMP5-KO cell lines compared to the parent cell lines (Fig. 3C).
Analysis of cancer-related protein and messenger RNA expression associated with LAMP5. a Analysis of alterations in cancer pathway-related protein expression after LAMP5 KO. Those upregulated more than 1.5-fold are listed above, and those downregulated less than 0.5-fold are listed below. b PCR array analysis of EMT-related mRNA expression. The genes associated with LAMP5 with a Spearman correlation coefficient greater than 5.0 are listed. c PCR array analysis of autophagy-related genes. Alterations after LAMP5 KO was analyzed. Genes upregulated more than twofold after LAMP5 KO are listed (left), and genes downregulated less than 0.5-fold are listed (right)
Protein expression analysis
Alterations in protein expression related to cancer progression between parent cells and LAMP5-KO cells were analysed using the Proteome profiler. ICAM-1/CD54, Serpin E1/PAI-1, EpCAM/TROP1, u-Plasminogen, Activator/Urokinase, CCL20/MIP-3α, Cathepsin S, CXCL8/IL-8, and CCL2/MCP-1 were increased more than 1.5-fold by LAMP5 KO. In contrast, Tenascin C, Kallikrein 6, VCAM-1/CD106 and endostatin were decreased less than 0.5-fold (Fig. 3A).
PCR array of EMT- and autophagy-related genes
The expression correlation between LAMP5 and EMT-related genes in GC cell lines was analysed by PCR arrays. The results showed that COL3A1, COL1A2, TIMP1, TMEM132A, ITGAV, ZEB1, MMP2 and VCAN expression levels were significantly correlated (P < 0.05) with the LAMP5 expression level (Fig. 3B). Analysis of autophagy-related gene expression alterations by LAMP5 KO revealed that GAA, CTSS, FAS, ATG10, WIPI1, HDAC6, MAP1LC3B, and TNFSF10 were increased more than twofold by LAMP5 KO, and IGF1, IFNG, RGS19, CXCR4, ESR1, EIF4G1, HSPA8 and FADD were decreased by less than 0.5-fold. Among them, ATG9B, SNCA, and IRGM increased more than fivefold, and IGF1 and IFNG decreased less than 0.1-fold (Fig. 3C).
Effect of LAMP5-KD in multiple GC cells
N87, NUGC3 and NUGC4 cells are selected for LAMP5-KD assays because they expressed relatively high levels of LAMP5 mRNA expression. q-PCR analysis revealed a successful knockdown of LAMP5 mRNA expression (Supplementary Fig. 2A). LAMP5-KD using the si-LAMP5-2 attenuated the proliferation of N87, NUGC3 and NUGC4 cells (Supplementary Fig. 2B).
In vivo analysis
A subcutaneous xenograft model revealed that the tumour size of the tumours derived from the KO-LAMP5-1 and KO-LAMP5-2 cells at 4 and 6 weeks after implantation was significantly smaller than that of the parent cells (Fig. 4A). In the peritoneal dissemination model, tumour progression of the tumours derived from the parent cells was detected with IVIS 2 weeks after dissemination, whereas the progression of the KO-LAMP5-1 and KO-LAMP5-2 cell-derived tumours was not detected (Fig. 4B). Analysis of the liver metastasis model showed that the KO-LAMP5-1 cells did not form liver metastases, and the KO-LAMP5-2 cells formed single and small liver metastases, although the parent cells formed multiple metastases (Fig. 4C).
Mouse xenograft model of MKN1 and two kinds of LAMP5-KO cells. a Percutaneous tumour model. Images of mice sacrificed 6 weeks after implantation and excised tumours (left) and quantification of tumour volumes after subcutaneous injection (right) are shown. b Peritoneal dissemination model. In vivo imaging of tumour growth and laparotomy findings 8 weeks after dissemination (left) and quantification of in vivo imaging 2, 4, 6, and 8 weeks after dissemination (right) are shown. c Portal vein injection model of liver metastasis. In vivo images 4, 8, and 12 weeks after tumour implantation are shown in the upper left, and laparotomy findings, macroscopic findings and formalin-fixed cross-section findings are shown in the upper right. Quantification of in vivo imaging 4, 8, and 12 weeks after implantation is shown in the lower graph
Analysis of clinical samples
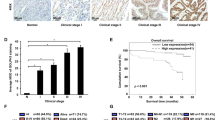
Clinical samples were analysed, and we defined patients whose cancer tissue LAMP5 mRNA expression was threefold higher than that in noncancerous tissue as the LAMP5 upregulated group. The percentages of peritoneal dissemination, positive peritoneal cytology, lymphatic involvement and nondifferentiation were significantly higher in the LAMP5-upregulated group than in the non-LAMP5-upregulated group (Table 2). The overall survival rate was significantly lower in the LAMP5-upregulated group (Fig. 5A).
Analysis of clinical samples. a Kaplan–Meier analysis of overall survival for patients with stage I–IV GC. Patients whose cancerous tissue showed threefold increases in LAMP5 mRNA expression compared to that of noncancerous tissue were defined as the LAMP5-upregulated group. b LAMP5 protein expression analysis of 17 GC patients with resected liver metastasis. Image of immunohistochemistry showing that liver metastatic tissues exhibited increased expression compared with primary GC tissues (L > S), equivalent expression (L = S) or reduced expression compared with primary GC tissues (L < S) (400 × magnification) (left). The distributions of immunostaining intensity are shown (right)
Immunohistochemistry
LAMP5 immunohistochemistry was performed on the GC non-cancerous stomach tissue, primary region and liver metastatic tissues of 17 patients who underwent gastrectomy and resection of liver metastasis. There were 12 cases in which metastatic region LAMP5 protein expression was higher than that of the primary region, 1 case of equal expression, and 4 cases in which metastatic region expression was lower than that of the primary region. The immunostaining images of each representative case and the distribution of immunostaining intensity are shown in Fig. 5B.
Discussion
LAMP5 is a single-transmembrane protein containing 280 amino acids, and its molecular weight is 31.7 kDa. LAMP5 harbors a luminal domain with four well-conserved cysteines, a transmembrane domain and a cytoplasmic tail that shares sequence and structural homology with LAMP1 and LAMP2[19]. LAMP family proteins are reported to have lysosomal functions and related autophagosomal and phagosomal functions[20]. LAMP5 was reported to be associated with the proliferation of plasmacytoid dendritic cells and related to the immune tolerance system, which is critical for tumorigenesis and tumour progression[19]. High LAMP5 expression was reported to be associated with poor prognosis of MLL, and knockdown of LAMP5 caused degradation of cancer-associated proteins through autophagy and improved the survival rate in vivo[20]. Concerning the association of LAMP5 with cancer, LAMP5 was reported to be part of a prognostic panel including 9 cancer-related genes in GC[21] and a prognostic biomarker of colorectal cancer [22]. As described above, the association of LAMP5 with GC progression through autophagy and macrophage invasion was predicted; however, there are no articles examining the role of LAMP5 in GC. Therefore, LAMP5 is considered to be a novel gene in the field of GC.
Cell cycle and apoptosis were analyzed to reveal the cause of the inhibited proliferation by LAMP5 KO in GC cells. The proportion of cells in G2 phase was increased by LAMP5 KO; therefore, G2 arrest was caused by LAMP5 KO, and LAMP5 is considered to regulate the G2 phase to promote cell cycle progression [23]. Total caspase activation and caspase 3, which is an effector caspase located downstream of the caspase cascade, were increased by LAMP5 KO; therefore, LAMP5 is considered to inhibit caspase and promote the progression of GC[24]. Moreover, the association of LAMP5 with the mitochondrial function was demonstrated because the expression of the initiator caspase 9, which is related to mitochondrial apoptosis and mitochondrial depolarization, was increased by LAMP5 KO[25]. Functional analysis of LAMP5-KO cells revealed that LAMP5 promotes cell functions such as invasion and migration to promote the distant metastasis of gastric cancer. Increasing ROS activation was reported to inhibit the formation of metastasis and is related to autophagy, EMT and cancer microenvironments [26, 27]. LAMP5 is considered to control ROS activation to promote the formation of metastasis. Recently, cancer stemness has been reported to have an important role in metastasis formation of malignant tumours[28]. LAMP5 is considered to promote metastasis, activating cancer stemness because activation of the cancer stemness marker ALDH was reduced and spheroid formation, which reflects the stemness potential[29], was inhibited by LAMP5 KO.
Subsequently, analysis of a mouse model of metastasis was performed to investigate whether LAMP5 activates metastasis in vivo. As a result, the suppressive effect of LAMP5 KO was stronger in the mouse metastasis models than on in vitro cell functions. Therefore, LAMP5 is considered to be closely involved in metastasis formation of GC. These results may lead to the development of drugs targeting LAMP5 because LAMP5 inhibition may prevent both hematogenous metastasis and peritoneal dissemination.
PCR array and protein expression analysis were performed to elucidate the molecular biological mechanisms by which LAMP5 promotes the metastasis of GC. EMT is an essential factor for the initial phase of metastasis formation[8], and various EMT-related molecules were positively correlated with LAMP5. COL1A2 and COL3A1 are extracellular matrix collagens reported to be related to EMT and macrophage invasion[30, 31]. MMP2 is involved in collagen metabolism and is reported to be related to metastasis formation in various kinds of cancer[32, 33]. ITGAV has been reported to be associated with the metastasis of many kinds of cancer and promote EMT with TGFB1 and collagen I by activating the PI3K cascade[34, 35]. ZEB1 is a transcription factor, and TMEM132A and VCAM are transmembrane proteins that are reported to enhance EMT[36, 37].
From the results of protein expression analysis, Tenascin, Kallikrein 6 and VCAM-1, which were downregulated by less than 0.5-fold by LAMP5 KO, were shown to be associated with cancer progression, macrophage invasion and autophagy[37,38,39]. In contrast, CCL2, CXCL8, and CCL20 were upregulated by more than twofold by LAMP5 KO and were also reported to be associated with macrophage invasion and tumour progression[40,41,42]. Therefore, LAMP5 was predicted to be involved in autophagy and macrophage invasion, as previously reported[20].
Thus, autophagy-related gene expression analysis was performed using LAMP5-KO cells to uncover the molecules associated with autophagy. Among the genes downregulated by LAMP5 KO, IGF1 was reported to be associated with the progression of colorectal cancer and ovarian cancer and to regulate autophagy[43]. CXCR4 is a component of many kinds of cancer pathways, such as the mTOR pathway, and is related to cancer proliferation, metastasis and chemoresistance, controlling autophagy and apoptosis [44]. LAMP5 may interact with these genes and promote metastasis of GC, modulating autophagy. Moreover, among the genes upregulated by LAMP5 KO, ATG9B was reported to show low expression in hepatic cancer and suppress proliferation, controlling autophagy, and it was reported to control the interaction of autophagy with LAMP1 in cervical cancer [45, 46]. IRGM and SNCA also associate with autophagy by interacting with the ATG family [47] and LAMP2, respectively [48]. LAMP5 is thought to suppress these molecules to promote metastasis of GC. LAMP5-KD resulted in a reduction of the proliferative capacity of the differentiated GC cell line (N87) and the undifferentiated GC cell lines (NUGC3 and NUGC4), suggesting that inhibition of LAMP5 was associated with a decreased aggressiveness across various types of GC cells. From the results of clinical samples, LAMP5 association with metastasis in GC was estimated because the percentages of peritoneal dissemination, positive peritoneal cytology, the undifferentiated type and vascular invasion in cancer metastasis were higher in the LAMP5-upregulated group than the non-upregulated group. LAMP5 may be applied in the clinical setting because LAMP5 expression predicts metastasis and poor prognosis of GC. Specifically, patients with high LAMP5 expression should receive PET and MRI to diagnose distant metastasis and stronger postoperative chemotherapy to prevent metastasis [49, 50].
This study has several limitations. First, there were no precise data on the association between LAMP5 expression in metastatic tissue and prognosis because there were few patients who underwent metastatic site resection for GC. Multicenter studies of cases with metastatic site resection will be needed to show the relationship between LAMP5 and metastasis in GC in clinical data. Second, further analysis of pathways that potentially interact with LAMP5 is needed to uncover the biological functions of LAMP5 in GC. Finally, metastatic suppression experiments should be performed using LAMP5 inhibitors in vivo to promote drug development.
In conclusion, LAMP5 is involved in various cell functions and plays a vital role in metastasis formation of GC. LAMP5 may be a promising novel target of drug development for metastatic GC in the future.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Kanda M, Shimizu D, Sawaki K, Nakamura S, Umeda S, Miwa T, et al. Therapeutic monoclonal antibody targeting of neuronal pentraxin receptor to control metastasis in gastric cancer. Mol Cancer. 2020;19:131.
Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–64.
Kanda M, Kasahara Y, Shimizu D, Miwa T, Umeda S, Sawaki K, et al. Amido-bridged nucleic acid-modified antisense oligonucleotides targeting SYT13 to treat peritoneal metastasis of gastric cancer. Mol Ther Nucleic Acids. 2020;22:791–802.
Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376:687–97.
Kang YK, Boku N, Satoh T, Ryu MH, Chao Y, Kato K, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–71.
Wilke H, Muro K, Van Cutsem E, Oh SC, Bodoky G, Shimada Y, et al. Ramucirumab plus paclitaxel versus placebo plus paclitaxel in patients with previously treated advanced gastric or gastro-oesophageal junction adenocarcinoma (RAINBOW): a double-blind, randomised phase 3 trial. Lancet Oncol. 2014;15:1224–35.
Huang L, Wu RL, Xu AM. Epithelial-mesenchymal transition in gastric cancer. Am J Transl Res. 2015;7:2141–58.
Massague J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529:298–306.
Brodt P. Role of the microenvironment in liver metastasis: from pre- to prometastatic niches. Clin Cancer Res. 2016;22:5971–82.
Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–95.
Stoecklein NH, Klein CA. Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int J Cancer. 2010;126:589–98.
Kanda M, Tanaka H, Shimizu D, Miwa T, Umeda S, Tanaka C, et al. SYT7 acts as a driver of hepatic metastasis formation of gastric cancer cells. Oncogene. 2018;37:5355–66.
Umeda S, Kanda M, Miwa T, Tanaka H, Tanaka C, Kobayashi D, et al. Expression of sushi domain containing two reflects the malignant potential of gastric cancer. Cancer Med. 2018;7:5194–204.
Umeda S, Kanda M, Miwa T, Tanaka H, Tanaka C, Kobayashi D, et al. Fraser extracellular matrix complex subunit 1 promotes liver metastasis of gastric cancer. Int J Cancer. 2020;146:2865–76.
Miwa T, Kanda M, Umeda S, Tanaka H, Shimizu D, Tanaka C, et al. Establishment of peritoneal and hepatic metastasis mouse xenograft models using gastric cancer cell lines. In Vivo. 2019;33:1785–92.
Kanda M, Shimizu D, Tanaka H, Tanaka C, Kobayashi D, Hayashi M, et al. Significance of SYT8 for the detection, prediction, and treatment of peritoneal metastasis from gastric cancer. Ann Surg. 2018;267:495–503.
Miwa T, Kanda M, Shimizu D, Umeda S, Sawaki K, Tanaka H, et al. Hepatic metastasis of gastric cancer is associated with enhanced expression of ethanolamine kinase 2 via the p53-Bcl-2 intrinsic apoptosis pathway. Br J Cancer. 2021;124:1449–60.
Combes A, Camosseto V, N’Guessan P, Arguello RJ, Mussard J, Caux C, et al. BAD-LAMP controls TLR9 trafficking and signalling in human plasmacytoid dendritic cells. Nat Commun. 2017;8:913.
Wang WT, Han C, Sun YM, Chen ZH, Fang K, Huang W, et al. Activation of the lysosome-associated membrane protein LAMP5 by DOT1L serves as a bodyguard for MLL fusion oncoproteins to evade degradation in leukemia. Clin Cancer Res. 2019;25:2795–808.
Wang Z, Chen G, Wang Q, Lu W, Xu M. Identification and validation of a prognostic 9-genes expression signature for gastric cancer. Oncotarget. 2017;8:73826–36.
Martinez-Romero J, Bueno-Fortes S, Martin-Merino M, Ramirez de Molina A, De Las-Rivas J. Survival marker genes of colorectal cancer derived from consistent transcriptomic profiling. BMC Genom. 2018;19:857.
Gire V, Dulic V. Senescence from G2 arrest, revisited. Cell Cycle. 2015;14:297–304.
Mazumder S, Plesca D, Almasan A. Caspase-3 activation is a critical determinant of genotoxic stress-induced apoptosis. Methods Mol Biol. 2008;414:13–21.
Pradelli LA, Beneteau M, Ricci JE. Mitochondrial control of caspase-dependent and -independent cell death. Cell Mol Life Sci. 2010;67:1589–97.
Poillet-Perez L, Despouy G, Delage-Mourroux R, Boyer-Guittaut M. Interplay between ROS and autophagy in cancer cells, from tumor initiation to cancer therapy. Redox Biol. 2015;4:184–92.
Sinha K, Das J, Pal PB, Sil PC. Oxidative stress: the mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch Toxicol. 2013;87:1157–80.
Triana-Martínez F, Loza MI, Domínguez E. Beyond tumor suppression: senescence in cancer stemness and tumor dormancy. Cells. 2020;9:346.
Lin RZ, Chang HY. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol J. 2008;3:1172–84.
Su B, Zhao W, Shi B, Zhang Z, Yu X, Xie F, et al. Let-7d suppresses growth, metastasis, and tumor macrophage infiltration in renal cell carcinoma by targeting COL3A1 and CCL7. Mol Cancer. 2014;13:206.
Yuan L, Shu B, Chen L, Qian K, Wang Y, Qian G, et al. Overexpression of COL3A1 confers a poor prognosis in human bladder cancer identified by co-expression analysis. Oncotarget. 2017;8:70508–20.
Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D, et al. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14:1365–80.
Huang Q, Lan F, Wang X, Yu Y, Ouyang X, Zheng F, et al. IL-1β-induced activation of p38 promotes metastasis in gastric adenocarcinoma via upregulation of AP-1/c-fos, MMP2 and MMP9. Mol Cancer. 2014;13:18.
Wang H, Chen H, Jiang Z, Lin Y, Wang X, Xiang J, et al. Integrin subunit alpha V promotes growth, migration, and invasion of gastric cancer cells. Pathol Res Pract. 2019;215:152531.
Feldkoren B, Hutchinson R, Rapoport Y, Mahajan A, Margulis V. Integrin signaling potentiates transforming growth factor-beta 1 (TGF-beta1) dependent down-regulation of E-Cadherin expression—important implications for epithelial to mesenchymal transition (EMT) in renal cell carcinoma. Exp Cell Res. 2017;355:57–66.
Oh-hashi K, Imai K, Koga H, Hirata Y, Kiuchi K. Knockdown of transmembrane protein 132A by RNA interference facilitates serum starvation-induced cell death in Neuro2a cells. Mol Cell Biochem. 2010;342:117–23.
Chen Q, Zhang XH, Massague J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 2011;20:538–49.
Yang ZT, Yeo SY, Yin YX, Lin ZH, Lee HM, Xuan YH, et al. Tenascin-C, a prognostic determinant of esophageal squamous cell carcinoma. PLoS ONE. 2016;11:e0145807.
Kim TW, Lee SJ, Kim JT, Kim SJ, Min JK, Bae KH, et al. Kallikrein-related peptidase 6 induces chemotherapeutic resistance by attenuating auranofin-induced cell death through activation of autophagy in gastric cancer. Oncotarget. 2016;7:85332–48.
Roca H, Varsos ZS, Mizutani K, Pienta KJ. CCL2, survivin and autophagy: new links with implications in human cancer. Autophagy. 2008;4:969–71.
Lin C, He H, Liu H, Li R, Chen Y, Qi Y, et al. Tumour-associated macrophages-derived CXCL8 determines immune evasion through autonomous PD-L1 expression in gastric cancer. Gut. 2019;68:1764–73.
Kadomoto S, Izumi K, Hiratsuka K, Nakano T, Naito R, Makino T, et al. Tumor-associated macrophages induce migration of renal cell carcinoma cells via activation of the CCL20-CCR6 axis. Cancers (Basel). 2019;12:89.
Liu L, Wang X, Li X, Wu X, Tang M, Wang X. Upregulation of IGF1 by tumor-associated macrophages promotes the proliferation and migration of epithelial ovarian cancer cells. Oncol Rep. 2018;39:818–26.
Teicher BA, Fricker SP. CXCL12 (SDF-1)/CXCR4 pathway in cancer. Clin Cancer Res. 2010;16:2927–31.
Tingting C, Shizhou Y, Songfa Z, Junfen X, Weiguo L, Xiaodong C, et al. Human papillomavirus 16E6/E7 activates autophagy via Atg9B and LAMP1 in cervical cancer cells. Cancer Med. 2019;8:4404–16.
Wang N, Tan HY, Li S, Feng Y. Atg9b deficiency suppresses autophagy and potentiates endoplasmic reticulum stress-associated hepatocyte apoptosis in hepatocarcinogenesis. Theranostics. 2017;7:2325–38.
Kumar S, Jain A, Choi SW, da Silva GPD, Allers L, Mudd MH, et al. Mammalian Atg8 proteins and the autophagy factor IRGM control mTOR and TFEB at a regulatory node critical for responses to pathogens. Nat Cell Biol. 2020;22:973–85.
Poehler AM, Xiang W, Spitzer P, May VE, Meixner H, Rockenstein E, et al. Autophagy modulates SNCA/alpha-synuclein release, thereby generating a hostile microenvironment. Autophagy. 2014;10:2171–92.
Kanda M, Suh YS, Park DJ, Tanaka C, Ahn SH, Kong SH, et al. Serum levels of ANOS1 serve as a diagnostic biomarker of gastric cancer: a prospective multicenter observational study. Gastric Cancer. 2020;23:203–11.
Nakanishi K, Kanda M, Umeda S, Tanaka C, Kobayashi D, Hayashi M, et al. The levels of SYT13 and CEA mRNAs in peritoneal lavages predict the peritoneal recurrence of gastric cancer. Gastric Cancer. 2019;22:1143–52.
Acknowledgements
We thank Edanz Group (http://www.edanzediting.com/ac) for editing a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
All procedures performed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. Nagoya University’s ethics committee approved this study (Approval number 2014-0043). All institutional and national guidelines for the care and use of laboratory animals were followed and were approved by Nagoya University Animal Research Committee (approval number 30143).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Umeda, S., Kanda, M., Shimizu, D. et al. Lysosomal-associated membrane protein family member 5 promotes the metastatic potential of gastric cancer cells. Gastric Cancer 25, 558–572 (2022). https://doi.org/10.1007/s10120-022-01284-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-022-01284-y