Abstract
Background
YAP1 is a core protein of the Hippo signaling pathway and is associated with malignancy and immunosuppression. In the present study, we discovered a novel lncRNA, RP11-323N12.5, with tumor promotion and immunosuppression activities through enhancing transcription of YAP1.
Methods
RP11-323N12.5 was identified using GEPIA. Its expression levels and their relationship with clinical features were investigated using clinical samples. The regulation of YAP1 transcription by RP11-323N12.5 was investigated in both GC and T cells, the tumor and immunosuppression promotion roles of RP11-323N12.5 were explored in vitro and in vivo.
Results
RP11-323N12.5 was the most up-regulated lncRNA in human GC, based on data from the TCGA database. Its transcription was significantly positively correlated with YAP1 transcription, YAP1 downstream gene expression which contribute to tumor growth and immunosuppression. RP11-323N12.5 promoted YAP1 transcription by binding to c-MYC in the YAP1 promoter region. Meanwhile, transcription of RP11-323N12.5 was also regulated by YAP1/TAZ/TEADs activation in GC cells. RP11-323N12.5 had tumor- and immnosuppression-promoting effects by enhancing YAP1 downstream genes in GC cells. Excessive RP11-323N12.5 was also observed in tumor-infiltrating leukocytes (TILs), which may be exosome-derived and also be related to enhanced Treg differentiation as a result YAP1 up-regulation. Moreover, RP11-323N12.5 promoted tumor growth and immunosuppression via YAP1 up-regulation in vivo.
Conclusions
RP11-323N12.5 was the most up-regulated lncRNA in human GC and it promoted YAP1 transcription by binding to c-MYC within the YAP1 promoter in both GC and T cells. RP11-323N12.5 is an ideal therapeutic target in human GC due to its tumor-promoting and immunosuppression characteristics.
Similar content being viewed by others
Background
Long noncoding RNAs (lncRNAs) have gained widespread attention by biological and medical researchers recently. However, decades ago, lncRNAs were termed “junk DNA” due to its transposons, pseudogenes, and simple repeats, which make up approximately 70% of the human genome [1, 2]. With the evolution of high-throughput RNA sequencing and genetic modification techniques, more functional lncRNAs have been investigated in various human diseases, especially in malignancies, such as lung cancer, gastric cancer, liver cancer, colorectal cancer, and leukemia [3,4,5,6]. The functional mechanisms of lncRNAs are varied and include chromatin remodeling and chromatin interactions, as well as acting as enhancers, promoters, or introns of other genes; pseudogenes; or natural antisense transcripts [7, 8]. In addition, lncRNAs may also host one or more small RNAs within their transcriptional units. Besides their function within tumor cells, tumor-derived lncRNAs also modulate the tumor microenvironment via exosomes, switching tumor immunity from anti-cancer to pro-cancer by targeting signaling pathways in different immunocytes [9,10,11,12].
Hippo signaling is one of the tumor- and tumor immunosuppression-related signaling pathways that was first discovered in Drosophila melanogaster through genetic mosaic screens [13, 14]. The Hippo signaling pathway consists of the NDR family protein kinase, Warts (Wts); the WW domain–containing protein, Salvador (Sav); the Ste20-like protein kinase, Hippo (Hpo); and the adaptor protein Mob as tumor suppressor (Mats) [13, 14]. Dysregulation of Hippo pathway components has been demonstrated in many cancers [15]. Besides its promotion of cell proliferation, dysregulation of Hippo signaling has also been shown to regulate many normal biological and disease processes, including survival, metabolism, stemness, angiogenesis, invasion, and immunosuppression [15, 16]. In addition to classical stimuli of Hippo signaling, many studies have reported its regulation by lncRNAs [17, 18]. Therefore, an investigation of the diagnostic value and mechanisms of lncRNAs that target Hippo signaling is warranted.
GEPIA is an online tool for analyzing RNA sequencing data from 9736 tumors and 8587 normal samples from the Cancer Genome Atlas Program (TCGA) and the Genotype-Tissue Expression (GTEx) project, using a standard processing pipeline [19]. Using this tool, we obtained 104 most differentially expressed transcripts between human gastric cancer and normal stomach tissues. Among these transcripts, we found that RP11-323N12.5 was the most significantly up-regulated lncRNA. Correlation analysis indicated that it was related to tumor growth and immunosuppression genes within the Hippo signaling pathway. In the present study, the clinical diagnostic value and functional mechanisms of RP11-323N12.5 were explored.
Materials and methods
TCGA database analysis and bioinformatics analysis
We used GEPIA, an online TCGA and GTEx analysis tool, to identify differentially expressed genes in the stomach adenocarcinoma (STAD) subset. TRANSFAC (www.gene-regulation.com) and PROM (https://alggen.lsi.upc.es/) were used to predict the potential binding sites in the promoter regions of YAP1 or RP11-323N12.5 (1000 bp upstream flanking sequence from the transcription start site).
Patients
This hospital-based case–control study included 67 GC patients. All subjects were recruited from the First People’s Hospital of Kunshan, affiliated with Jiangsu University, between January 2017 and December 2018. All patients underwent surgery for primary GC and subjects with a previous history of cancer or chemotherapy were excluded. This study was approved by the Ethics Review Board of the First People’s Hospital of Kunshan (KSL-2017017) and all patients provided written informed consent.
Cell lines and reagents
Human GC cell lines, including MKN45 and MGC-803, were purchased from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco, Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA) and maintained at 37 °C in a humidified atmosphere containing 5% CO2. Lysophosphatidic acid (LPA) was purchased from ENZO (BML-LP100-0025; Farmingdale, NY, USA) and an shRNA targeting YAP was purchased from Santa Cruz Biotechnology (sc-38637-V; Dallas, TX, USA). Positive clones were obtained using a puromycin screen.
Real-time PCR
Total RNA was isolated from GC tissues and cell lines using TRIzol reagent (Invitrogen, 15596026). Gene expression was assessed using a SYBR green-based real-time PCR method (AceQ Universal SYBR qPCR Master Mix, Vazyme, Nanjing, China, Q511-02). The primers used in this study are listed in Supplementary Table 1.
Multicolor immunohistochemistry, immunofluorescence, and in situ hybridization
For multicolor staining, we processed slides strictly according to the instructions of the Opal 4-Anti-Rabbit Automation IHC kit (NEL830001KT; AKOYA Biosciences, Marlborough, MA, USA). Slides were incubated at 4 °C overnight with primary anti-YAP1 (#4912; Cell Signaling Technology, Danvers, MA, USA), anti-Survivin (ab76424; Abcam, Cambridge, UK) anti-human CD4 (ab181724, Abcam), anti-mouse CD4 (ab183685, Abcam), anti-COX2 (ab15191, Abcam), anti-human Foxp3 (ab20034, Abcam), and anti-mouse Foxp3 antibodies (ab75763, Abcam) and then photographed using a digital microscope camera (Nikon, Tokyo, Japan).
In situ hybridization (ISH) was performed using a Boster ISH kit (Boster, Wuhan, China). Cells were fixed and permeabilized using methanol and protease to allow biotin-labeled probes to access the cells. Cells were treated with 30% H2O2 in ddH2O, at a ratio of 1:10, for 5 min, after which pepsase, diluted in 3% citric acid, was applied for 20 s. The second fixation was performed using 1% paraformaldehyde/0.1 M PBS. Next, the cells were incubated with pre-hybridization solution at 40 °C for 2 h and then with lncRNA target probes at 30 °C overnight, followed by two washes with 2× saline sodium citrate. After blocking, a biotin-labeled anti-digoxin antibody was added and cells were incubated for 60 min. The following sequence was used for the RP11-323N12.5 probe: ctcctgcttggggggcgccgcgccgc.
Western blotting
For western blotting, proteins were extracted from cultured cells using RIPA buffer containing phenylmethanesulfonylfluoride (Beyotime, Nantong, China). An equal amount of protein per sample (100 μg) was separated by 7.5%/12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. The following primary polyclonal antibodies were used: anti-YAP1 (#4912, Cell Signaling Technology), anti-phospho(S127)-YAP1 (#13008, Cell Signaling Technology), anti-Survivin (ab76424, Abcam), anti-MMP9 (ab219372, Abcam), and anti-COX2 (ab15191, Abcam). HRP-conjugated anti-rabbit secondary antibodies were used (Abcam). The blots were developed using ECL reagent (WBKLS0500; Merck Millipore, Burlington, MA, USA). An equal amount of protein loading in each lane was confirmed using an anti-β-Actin antibody (ab8226, Abcam). ImageJ software was used to quantify the integrated density of the protein bands.
Luciferase gene reporter assay
The promoter region of the YAP1 gene containing wild-type (WT) or mutant (MU) potential target sites for c-MYC, as well as the promoter region of the RP11-323N12.5 gene containing wild-type or mutant binding sites for TEAD1, were designed and synthesized by Genescript, Co. (Nanjing, China) and inserted into the pGL4 Vector (Promega, Madison, WI, USA). For luciferase assays, MKN45, MGC803, and Jurkat cells were transfected with pGL4-YAP1 WT pro or pGL4-YAP1 c-MYC MU pro, pGL4-RP11-323N12.5 WT pro or pGL-4-RP11-323N12.5 TEAD1 MU pro, or control vector, using Lipofectamine 2000 (11668019, ThermoFisher Scientific, Waltham, MA, USA). LPA-treated cells were harvested 48 h after transfection and a luciferase assay was performed using a Dual-Luciferase® Reporter Assay System (E1910, Promega, Madison WI,), according to the manufacturer’s protocol.
Chromatin isolation by RNA purification assay (ChIRP)
Chromatin isolation by RNA purification (ChIRP) assays were performed using an EZ- Magna ChIRP RNA Interactome Kit (17-10495, Millipore). The sequence of the RP11-323N12.5 probe was GGGCGTTGGAGGCCTGC. Isolated RNA was used for qRT-PCR analysis to quantify the enrichment of chromatin. The qRT-PCR primer sequences used in this study were as follows: YAP1 promoter: 5′-AATGAGTTTCACTGAGGGAT-3′ (forward) and 5′-CAAAACAAGCAATGTGAGGT-3′ (reverse).
CCK8 cell proliferation, cell colony formation, migration, and invasion assays
A CCK-8 assay kit (CK04-11, Dojindo, Kumamoto, Japan) was used to detect cell proliferation, according to the manufacturer’s protocol. Cells were seeded into 96-well plates (500 cells/well in 100 μL of medium) and 10 μL of CCK-8 solution was added to each well at the same time each day for each time-point. After incubation for 2 h at 37 °C, the absorbance (450 nm) of each well was measured.
For cell colony formation assays, GC cells were plated in 6-well plates, at a density of 500 cells/well, and incubated for 2 weeks. Proliferating colonies were fixed with 75% alcohol and stained with crystal violet. Colonies containing ≥ 50 cells were counted under a microscope. All procedures were performed in triplicate.
For migration and invasion assays, cells were placed into the upper chamber of a transwell plate (8-μm pore size; Millipore), which was coated with (invasion assay) or without (migration assay) a Matrigel mix (BD Biosciences, San Jose, CA, USA), according to the manufacturer’s protocol. After incubation for 24 h, the cells located on the upper membrane of the transwell chambers were removed with cotton wool, while cells that had migrated or invaded through the membrane were fixed with methanol for 10 min and stained with 0.1% crystal violet for 30 min at 37 °C. Cells in three randomly selected microscopic fields were counted for each group. Experiments were independently repeated three times.
ELISA
The secretion of PGE2 and CXCL15 into the supernatant of MKN45 and MGC-803 cells, RP11-323N12.5-overexpressing cells, and RP11-323N12.5 + YAP1 shRNA-transfected cells was measured using ELISA kits (ab133021 for PGE2, ab234567 for CXCL15; Abcam).
Treg cell differentiation and flow cytometric analysis
Various genetically modified Jurkat cells were obtained for the in vitro stimulation of Treg cell polarization. For the in vitro generation of Treg cells, Jurkat cells were stimulated with recombined human TGF-β (1 ng/mL; Peprotech, Rocky Hill, NJ, USA) for 7 days. Dead cells were excluded based on staining with a live/dead fixable dye. Thereafter, cells were fixed, permeabilized with IntraPrep reagent (BD Pharmingen, San Jose, CA, USA), and then stained with fluorochrome-conjugated antibodies against CD4 (20 μL per test, 555346), CD25 (20 μL per test, 555432), or Foxp3 (20 μL per test, 560082, BD Pharmingen). Data were acquired on a BD FACSVerse™ flow cytometer (BD Pharmingen).
Tumor-infiltrating leukocyte isolation
Fresh tissue–infiltrating lymphocytes were obtained as described previously [20]. Briefly, GC tissue specimens were cut into small pieces and digested in RPMI 1640. Dissociated cells were filtered through a 75 μm cell strainer and separated by Ficoll centrifugation. The mononuclear cells were then washed and resuspended in RPMI 1640, supplemented with 10% FBS (Gibco, Waltham, MA, USA). T cells were purified using anti-CD3 magnetic Dynabeads (Invitrogen Life Technologies, CA, USA) according to manufacturer’s instruction.
Subcutaneous tumorigenesis assay
For subcutaneous tumorigenesis assays, tumor cells were injected subcutaneously in mice (2 × 106 cells/100 μl per flank containing 1 × 106 GC tumor cells and 1 × 106 peripheral mono-nuclear blood cells obtained from non-GC healthy volunteers) [20]. Four weeks after injection, the mice were euthanized. The tumor volume was calculated using the formula (width2 × length)/2. The tumor tissues were then harvested for further analysis.
Statistical analysis
Data are presented as mean ± SD. Chi square tests and a two-tailed Student’s t test analysis of variances were used to evaluate statistical differences in demographic and clinical characteristics. All in vitro expression experiments were repeated at least three times, each with triplicate samples. Pearson’s correlation analysis was used to analyze the relationship between associated factors. Statistical analyses were performed using STATA 9.2 (StataCorp, College Station, TX, USA) and presented using Prism software (GraphPad, San Diego, CA, USA). In all cases, P < 0.05 was considered significant.
Results
RP11-323N12.5 is the most overexpressed lncRNA in human gastric cancer, based on the TCGA data base
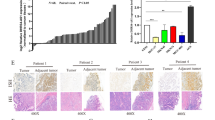
Based on the newly developed interactive web server, GEPIA [19], we found 3746 significantly up-regulated genes in stomach adenocarcinomas (STADs) compared to their paired normal tissues (n = 36) using an ANOVA to analyze TCGA data. The chromosomal distribution of the top 104 differentially expressed genes is presented in Fig. 1a. The top 25 up-regulated genes are listed in Fig. 1b. RP11-323N12.5 was found to be the most significantly up-regulated lncRNA in human GC tissues (Gene ID ENSG00000267601.1), with a median expression of 27.970 in tumors and 0.000 in normal tissues, with an adjusted P value 1.36e-67 (Fig. 1b). RP11-323N12.5 is an anti-sense non-coding RNA located on chr17:78855478–78855844, with a transcript of 367 bp. Moreover, using a one-way ANOVA to analyze samples from the GTEx database, we found that RP11-323N12.5 was also overexpressed in human GC tissues compared to normal tissues (GC n = 408 vs. normal n = 211, Fig. 1c). We found that RP11-323N12.5 expression levels were significantly higher in advanced GC (TMN Stage I–II vs. Stage III, IV; Fig. 1d). Survival analysis showed that high RP11-323N12.5 expression was significantly correlated with disease-free survival (P = 0.047), but not with overall survival of post-surgery GC patients (Fig. 1e).
RP11-323N12.5 was the most up-regulated lncRNA in human gastric cancer associated with immunosuppression in the TCGA database. a The chromosomal distribution of over-expressed genes in human gastric cancer (GC). b A list of the top 25 up-regulated genes in human GC. c Comparison of RP11-323N12.5 transcription in human GC (n = 408) vs. adjacent normal tissues (n = 211). d Transcription of RP11-323N12.5 in different stages of human GC. e Survival analysis evaluating the association of RP11-323N12.5 with disease-free survival of post-surgery GC patients. f–m Linear correlation analysis of RP11-323N12.5 expression and YAP1, AXL, TGFBR1, TGFB1, FOXP3, IL17A, IL4, or IFNg
Through pathway enrichment analysis between RP11-323N12.5-high and RP11-323N12.5-low GC samples (n = 408), we found that RP11-323N12.5 expression was significantly related to Hippo signaling (data not shown). Furthermore, we confirmed this correlation by investigating the correlation between RP11-323N12.5 and components of the Hippo signaling pathway and its downstream genes. These results indicated that RP11-323N12.5 expression levels were significantly positively correlated with YAP1, AXL, and TGFBR1 expression levels (Fig. 1f–h). Since, the inactivation of Hippo signaling is significantly associated with a suppressive tumor microenvironment (TME) in human solid tumors, we also investigated the correlation between RP11-323N12.5 expression and the expression of genes representing different types of T helper cells. We found that RP11-323N12.5 was significantly correlated with FOXP3 and TGFB1 expression (Fig. 1i, j), which implied that RP11-323N12.5 was associated with Treg cells, but not Th1, Th2, or Th17 cells in the TME of human GC (Fig. 1k–m).
RP11-323N12.5 was associated with YAP1 expression in human GC
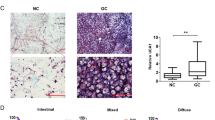
To confirm the overexpression of RP11-323N12.5 in human GC, we collected 67 paired human GC and para-tumor tissues. We found that RP11-323N12.5 was overexpressed in GC tissues compared to paired normal tissue (Fig. 2a). Moreover, RP11-323N12.5 expression levels were also significantly higher in advanced stage GC (stage III–IV) compared to early stage GC (stage I–II, Fig. 2b). The transcription levels of YAP1 were also determined by real-time PCR in 67 paired GC and normal tissues. Similar to previously published results [19], YAP1 was found to be overexpressed in GC tissues and its expression levels were positively correlated with RP11-323N12.5 expression levels (P = 0.0004, r = 0.6275). Moreover, we found that the downstream genes of Hippo signaling, BIRC5, AXL, and PTSG2 were overexpressed in GC tissues (Fig. 2c, d, e) and their expression levels were also positively correlated with RP11-323N12.5 expression levels in human GC tissues (BIRC5 vs. RP11-323N12.5: r = 0.7515, P < 0.0001; AXL vs. RP11-323N12.5: r = 0.3885, P = 0.0012; and PTSG2 vs. RP11-323N12.5: r = 0.5000, P = 0.0009; Fig. 2f, g, h).
RP11-323N12.5 was over-expressed in human GC tissues and was associated with the dysregulation of Hippo signaling. a Transcription of RP11-323N12.5 in paired human GC and adjacent normal stomach tissues (n = 67). b RP11-323N12.5 expression at different stages of human GC. c Transcription of YAP1 in paired human GC and adjacent normal stomach tissues (n = 67). d Linear correlation analysis of RP11-323N12.5 and YAP1 in human GC. c–e Transcription of BIRC5, AXL, and PTSG2 in paired human GC and adjacent normal stomach tissues (n = 67). f–h Linear correlation analysis of RP11-323N12.5 and BIRC5, AXL, or PTSG2 in human GC. i Representative figures of immunofluorescence (IF) staining of YAP1 and Survivin in human GC tissues with high and low levels of RP11-323N12.5 expression. j, k Integrated optical intensity (IOD) analysis of YAP1 and Survivin protein expression in human GC tissues with high (n = 33) and low (n = 34) levels of RP11-323N12.5 expression. l, m Linear correlation analysis of RP11-323N12.5 and YAP1 or Survivin in human GC with high (n = 33) and low (n = 34) levels of RP11-323N12.5 expression. Each experiment was performed in triplicate. Data are presented as the mean ± SEM and were analyzed by Student’s t test (**P < 0.01)
We also detected the protein expression of YAP1 and the downstream, protein, Survivin, in human GC tissues by immunofluorescence staining, in two groups according to RP11-323N12.5 expression (RP11-323N12.5-high, n = 33 and RP11-323N12.5-low, n = 34). We found that YAP1 and Survivin levels were higher in the RP11-323N12.5-high group than in the RP11-323N12.5-low group (Fig. 2i, j, k). YAP1 and Survivin protein levels were also positively correlated with RP11-323N12.5 expression levels (RP11-323N12.5 vs. YAP1: P = 0.0001, r = 0.4535; RP11-323N12.5 vs. Survivin: P < 0.0001, r = 0.5079). Based on these data, we proposed that RP11-323N12.5 may be associated with YAP1 transcription and downstream gene activation.
RP11-323N12.5 and YAP1 promoted the transcription of each other by promoter binding in GC cells
Since clinical studies have indicated that RP11-323N12.5 expression is positively correlated with YAP1 transcription, we further investigated the detailed regulatory mechanisms of RP11-323N12.5 and YAP1. Firstly, we overexpressed and knocked down RP11-323N12.5 expression in the human GC cell lines, MKN45 and MCG803, respectively. We found that the changes in YAP1 transcription were synchronized to RP11-323N12.5 expression (Fig. 3a). We also investigated YAP1, which showed a similar trend to its transcription levels (Fig. 3b). However, the expression of RP11-323N12.5 had no effect on YAP1 activation. The YAP1 activator, LPA (0.1 µM) can decrease the phosphorylation of the S127 residue of YAP1 and increase total YAP1 protein levels, but RP11-323N12.5 had no effect on the ratio of YAP1(S127)/YAP1 (Fig. 3c). These results implied that RP11-323N12.5 can regulate YAP1 transcription, but not YAP1 activation.
RP11-323N12.5 and YAP1 promoted the transcription of each other by promoter binding in GC cells. a The transcription of RP11-323N12.5 and YAP1 in MKN45 and MGC-803 cells with different treatments. b Expression of YAP1 in MKN45 and MGC-803 cells with different treatments, using β-actin as an internal control. c Western blotting analysis of the levels of phosphorylated (S127) and total YAP1 in MKN45 cells after treatment with 0.1 µM LPA or DMSO (control) for 6 h. d Schematic diagram of the mechanism whereby RP11-323N12.5 promotes YAP1 transcription by binding to c-MYC in the YAP1 gene promoter. e Luciferase gene reporter assay of YAP1 promoter activity in MKN45 and MGC-803 cells with different treatments. f RNA retrieval rate in chromatin isolation by RNA purification (ChIRP) assays, using probes targeting RP11-323N12.5 and LacZ (control). g c-MYC and c-Jun protein detection in proteins obtained from the ChIRP assay. h Co-localization assay determining the distribution and expression of RP11-323N12.5 (by ISH) and c-MYC (by IF) in the human GC cell lines, MKN45 and MGC-803. i, j RP11-323N12.5 transcription was measured by real-time PCR and ISH in MKN45 and MGC-803 cells, 6 h after treatment with DMSO (control) or LPA (0.1 µM). k Schematic diagram of the promotion of RP11-323N12.5 transcription by YAP/TAZ/TEADs. l Luciferase gene reporter assay of RP11-323N12.5 promoter activity in MKN45 and MGC-803 cells treated with LPA (0.1 µm) or DMSO (1 h after treatment). Each experiment was performed in triplicate. Data are presented as the mean ± SEM and were analyzed by Student’s t test (**P < 0.01)
We screened for potential binding between RP11-323N12.5 and transcription factors (TFs) at the YAP1 gene promoter and found that c-Myc was the TF linking RP11-323N12.5 and YAP1 transcription (YAP1 promoter region [− 216 bp to − 211 bp], Fig. 3d). Based on this prediction, we constructed WT and c-Myc binding site mutation (c-Myc MU) YAP1 promoters and found that RP11-323N12.5 overexpression increased and RP11-323 N12.5 knockdown decreased YAP1 promoter activity. However, RP11-323N12.5 expression had almost no effect on the YAP1 promoter with the c-Myc binding site mutated (Fig. 3e). We also used a ChIRP assay to confirm the binding of RP11-323N12.5 to c-MYC. Specific primers targeting c-Myc bound to the promoter region retrieved a DNA sequence pull downed by the RP11-323N12.5 probe. Western blotting analysis further confirmed the existence of c-Myc protein in the RP11-323N12.5 probe precipitate (Fig. 3f, g). Lastly, we investigated the location of RP11-323N12.5 and c-Myc in MKN-45 and MGC 803 cells. ISH results indicated that RP11-323N12.5 was located mainly in the nucleus and we found that c-Myc and RP11-323N12.5 co-localized in the nuclei of both MKN-45 and MCG 803 cells (Fig. 3h).
Since RP11-323N12.5 was overexpressed in GC and its transcription was positively correlated with Hippo inactivation, based on our clinical investigation and data from the TCGA database, we next investigated whether YAP1 activation could promote RP11-323N12.5 transcription. Firstly, LAP-induced YAP1 activation was shown to significantly increase RP11-323N12.5 transcription in both MKN45 and MCG 803 cells, as determined by both real-time PCR and ISH analyses (Fig. 3i, j). When we queried the promoter region of RP11-323N12.5, we found that a TEAS1 binding site existed within this region (Fig. 3k). Therefore, we performed a luciferase gene reporter assay and found that LAP did not increase RP11-323N12.5 promoter activity when the TEAD1 binding site was mutated (Fig. 3l). These results indicated that RP11-323N12.5 and YAP1 can promote each other’s expression in human GC.
RP11-323N12.5 promoted GC cell proliferation and invasion, as well as immunosuppression, by increasing YAP1 expression
Since RP11-323N12.5 and YAP1 can promote each other’s expression in human GC, we investigated the mechanisms of this phenomenon in vitro. Three types of cell lines were constructed including WT cells; cells overexpressing RP11-323N12.5 (RP11-323N12.5); and cells overexpressing RP11-323N12.5, with YAP1 knocked down (RP11-323N12.5 + YAP1 shRNA). Cell proliferation was monitored using CCK8 assay and colony formation assays. We found that RP11-323N12.5 promoted the proliferation of both MKN45 and MGC803 cells; however, this effect was mainly dependent on YAP1 expression (Fig. 4a, b). RP11-323N12.5 markedly increased the migration and invasion ability of both GC cell lines. When YAP1 was knocked down by a specific shRNA, cell migration and invasion decreased to levels significantly less than those observed in WT cells (Fig. 4c). These data implied that RP11-323N12.5 is a YAP1-dependent oncogene. These oncogenic effects may be carried out by up-regulating Survivin, MMP9, and AXL, which were up-regulated by RP11-323N12.5 overexpression and down-regulated by knocking down YAP1 (Fig. 4d, e).
RP11-323N12.5 promoted cell proliferation, migration, and invasion by up-regulating YAP1. The human GC cell lines, MGC-803 and MKN45, underwent various treatments and cell proliferation (a), colony formation (b), and cell migration and invasion ability (c) were then determined. d Gene expression was measured by real-time PCR in MGC-803 and MKN45 cells with different treatments. All of the tested genes are downstream of YAP1 activation. e The protein downstreams of YAP1 activation were measured by western blotting in MGC-803 and MKN45 cells with different treatments. Each experiment was performed in triplicate. Data are presented as the mean ± SEM and were analyzed by Student’s t test (**P < 0.01)
Since previous studies have shown that inactivation of Hippo signaling is significantly related to immunosuppression [21, 22] and data from TCGA also implied that RP11-323N12.5 was significantly correlated with Treg cell-related immunosuppression, we analyzed genes downstream of YAP1 and immunosuppression-related genes, including PSTG2, CSF1, CXCL15, and CSF3. We found that RP11-323N12.5 significantly promoted the expression of these genes in a YAP1-dependent manner (Fig. 4d, e).
RP11-323N12.5 was related to Treg cell- and MDSC-related immunosuppression in the human GC tumor microenvironment
Based on in vitro studies, we found that RP11-323N12.5 may be related to Treg cell differentiation via cytokine secretion. We also investigated the relationship between RP11-323N12.5 expression and Treg cell distribution by multicolor immunohistochemistry (IHC). We found that in tissues with high levels of RP11-323N12.5 expression, the percentage of Treg cells was much higher than in tissues with low levels of RP11-323N12.5 expression. This was accompanied by higher levels of COX2 expression, which are reported to induce Tregs by increasing PGE2 expression in cancer (Fig. 5a–c). Moreover, we found a positive correlation between RP11-323N12.5 expression levels and the percentage of Tregs and COX2 expression in human GC (Fig. 5d, e). Next, we found more Tregs and MDSCs in GC tissues with high levels of RP11-323N12.5 expression than in those with low levels of RP11-323N12.5 expression (Figs. 5f, g, S1a, b). Furthermore, we found that serum PGE2 and CXCL15 levels were markedly higher in GC patients with high levels of RP11-323N12.5-expression (Fig. 5h, i).
RP11-323N12.5 was involved in Treg cell-related immunosuppression in the human GC tumor microenvironment. a Representative figures of multicolor staining of CD4, COX2, and FOXP3 in human GC tissues with high and low levels of RP11-323N12.5 expression. b, c Integrated optical density (IOD) values of COX2 and CD4 + FOXP3 + cell percentage (CD4 + FOXP3 +/CD4) in human GC tissues with high (n = 33) and low (n = 34) levels of RP11-323N12.5 expression. d, e Linear correlation analysis of RP11-323N12.5 and CD4 + FOXP3 percentage (log2) or COX2 IOD values (log2) in human GC. f, g Tumor-infiltrating leukocytes were isolated from human GC tissues with high (n = 33) and low (n = 34) RP11-323N12.5 expression levels and the percentages of Treg cells (CD4 + CD25 + FOXP3 +) and MDSCs (CD11b + CD33 +) were determined by flow cytometry. h, i The concentrations of PGE2 and Cxcl15 were determined in the serum of patients with high (n = 33) and low (n = 34) RP11-323N12.5 expression levels. j, k The levels of PGE2 and Cxcl15 secreted from MKN45 and MGC-803 cells after different treatments were determined by ELISA. l The human T cell isolated from six non-GC persons was co-cultured with MKN45 cells treated with TGF-β (1 ng/mL) for 7 days and the percentage of Treg cells (CD4 + CD25 + FOXP3 +) was determined by flow cytometry. Each experiment was performed in triplicate. Data are presented as the mean ± SEM and were analyzed by Student’s t test (**P < 0.01)
We next investigated PGE2 and CXCL15 secretion by human GC cell lines after various treatments. We found that RP11-323N12.5 promote the secretion of these proteins in a YAP1-dependent manner (Fig. 5j, k). We also co-cultured human T cells, isolated from six non-GC individuals, with MKN45 cells, after various treatments, for 7 days. We found that Treg cell differentiation was significantly increased by co-culture with RP11-323N12.5-overexpressing MKN45 cells, but this effect was inhibited by YAP1 depletion in MKN45 cells (Fig. 5l).
RP11-323N12.5 promoted Treg cell differentiation by enhancing YAP1 transcription in T cells
Since we observed that RP11-323N12.5 was related to Treg cell-induced immunosuppression by tumor-derived secretion, we next investigated whether endogenous RP11-323N12.5 could promote Treg cell differentiation. Firstly, we investigated YAP1 protein expression in CD4+ T cells using multicolor IHC. We found that T cell expression levels of YAP1 were higher in GC tissues expressing high levels of RP11-323N12.5 compared to those expressing low levels of RP11-323N12.5. Moreover, RP11-323N12.5 levels were also significantly positively corelated with the expression levels of these genes (Fig. 6c). These results indicated that endogenous RP11-323N12.5 in T cells was not only related to tumor cell RP11-323N12.5 expression, but also related to Treg cell differentiation. TILs were isolated from human GC tissues and RP11-323N12.5 levels were found to be significantly higher in GC tissues expressing high levels of RP11-323N12.5 than in those with low levels of RP11-323N12.5 expression. Treg cell-related genes, including YAP1, FOXP3, TGFBR2, and IL10 were all expressed at significantly higher levels in T cells isolated from tissues expressing high levels of RP11-323N12.5 compared to those expressing low levels of RP11-323N12.5. We wondered that RP11-323N12.5 whether can promote YAP1 transcription in T cells.
RP11-323N12.5 promoted Treg cell differentiation by enhancing YAP1 transcription in T cells. a Representative figures of multicolor staining of CD4, YAP1, and FOXP3 in human GC tissues with high and low levels of RP11-323N12.5 expression. b Integrated optical density (IOD) values of YAP1 expression in CD4+ T cells in human GC with high (n = 33) and low (n = 34) levels of RP11-323N12.5 expression. c Linear correlation analysis of YAP1 IOD values in CD4+ T cells and the percentage of CD4 + FOXP3 cells in human GC. d The transcription of RP11-323N12.5 was measured by real-time PCR in tumor-infiltrating lymphocytes (TILs) isolated from human GC tissues with high (n = 33) and low (n = 34) levels of RP11-323N12.5 expression. e The transcription of IL10, FOXP3, TGFBR2, and FR4 was determined in TILs isolated from human GC tissues with high (n = 33) and low (n = 34) levels of RP11-323N12.5 expression. f Jurkat cells with different levels of RP11-323N12.5 expression were treated with TGF-β 1 ng/mL for 7 days and the percentage of Treg cells (CD4 + CD25 + FOXP3) was determined by flow cytometry. g ChIRP assays were performed in Jurkat cells using probes targeting RP11-323N12.5 and LacZ (control). h Luciferase gene reporter assay of YAP1 gene promoter activity in Jurkat cells with different levels of RP11-323N12.5 expression. i Detection of RP11-323N12.5 in serum exosomes of GC patients with high (n = 33) and low (n = 34) levels RP11-323N12.5 expression. j, k The measurement of RP11-323N12.5 in supernatant exosomes of cultured MKN45 and MGC-803 human GC cell lines and Jurkat cells with RP11-323N12.5 knockdown. Each experiment was performed in triplicate. Data are presented as the mean ± SEM and were analyzed by Student’s t test (**P < 0.01)
We used Jurkat cells, a human T cell line, to further investigate this mechanism. Firstly, we found that overexpression of RP11-323N12.5 in Jurkat cells enhanced Treg cell differentiation and this effect was YAP1 activation dependent (Fig. 6f). Both mRNA and protein expression of YAP1 were up-regulated by RP11-323N12.5 (Fig. S2a, b). Next, we used a ChIRP assay to show that RP11-323N12.5 promoted YAP1 transcription by binding to c-MYC, specifically in the promoter region of the YAP1 gene (Fig. 6g). Similar results were seen in luciferase gene reporter assays, indicating that YAP1 promoter activity can be significantly increased by RP11-323N12.5 in a c-MYC binding-dependent manner, since promoter activity was fully restored once the c-MYC binding site was mutated (Fig. 6h). However, using both luciferase gene reporter assays and real-time PCR, we found that YAP1 activation had no effect on RP11-323N12.5 transcription in Jurkat cells (Fig. S2c, d). Next, we measured RP11-323N12.5 levels in both GC patient serum samples and in cell culture supernatants. We found that RP11-323N12.5 can be detected in the exosomes and its quantity varied according to RP11-323N12.5 expression levels within tumor cells (Fig. 6i, j, k). Furthermore, we co-cultured RP11-323N12.5 shRNA-transfected Jurkat cells with human GC cells showing different levels of RP11-323N12.5 expression. The result indicated that RP11-323N12.5 can transfer into Jurkat cells by exosome delivery (Figs. 6j, k, S2e). Therefore, we concluded that the high levels of RP11-323N12.5 expression in T cells were exosome derived.
RP11-323N12.5 promoted tumor growth and immunosuppression via YAP1 up-regulation in vivo
To investigate the pro-cancer and pro-immunosuppressive effects of RP11-323N12.5 in vivo, we performed a subcutaneous tumorigenesis assay using wild-type MKN45 cells; MKN45 cells overexpressing RP11-323N12.5; and MKN45 cells overexpressing RP11-323N12.5, with YAP1 knockdown. Simultaneously, the human mono-nuclear peripheral blood cells mainly containing T cells and monocytes were implanted subcutaneously with the tumor cells. RP11-323N12.5 significantly promoted tumor growth, but this effect was completely inhibited by YAP1 knockdown in vivo (Fig. 7a). The TME derived from different tumor cells was also investigated by multicolor IHC staining of YAP1, Foxp3, and Cd4. We found that YAP1 was up-regulated by RP11-323N12.5 in vivo and the number of infiltrating Treg cells increased in tumors derived from RP11-323N12.5-overexpressing MKN45 cells. However the number of infiltrating Treg cells decreased significantly when YAP1 was knocked down in MKN45-derived tumor tissue (Fig. 7b–d). Furthermore, TILs were isolated from the tumors and the numbers of both Treg cells and MDSCs were found to be increased in RP11-323N12.5-overexpressing tumors. However, their numbers decreased markedly when YAP1 expression was silenced in RP11-323N12.5-overexpressing tumor tissues (Fig. 7e, f).
RP11-323N12.5 promoted tumor growth and immunosuppression in vivo. a Subcutaneous tumorigenesis assay using MKN45 cells with different levels of gene expression, as indicated (n = 6 per group, left panel). Comparison of tumor volume in all three groups (right panel). b Multicolor staining of YAP1, CD4, and Foxp3 in tumor tissues. c, d Comparison of YAP1 expression and CD4 + Foxp3 + cells within tumor tissues of all three groups. e, f Comparison of Treg cells and MSDCs within tumor-infiltrating lymphocytes isolated from tumor tissues of all three groups. Each experiment was performed in triplicate. Data are presented as the mean ± SEM and were analyzed by Student’s t test (**P < 0.01)
In summary, in the present study, we used data from the TCGA database to identify RP11-323N12.5 as the most differentially expressed lncRNA between human GC and normal tissue. RP11-323N12.5 was shown to promote the transcriptional activation of YAP1 in GC cells and further promote immunosuppression by Treg cell differentiation and MDSCs infiltration via a YAP1 activation dependent manner (Fig. 8).
Discussion
In the present study, we found that a novel lncRNA, RP11-323N12.5, contributed to the overexpression of YAP1 in human GC. Although it is a controversial notion that YAP overexpression alone is related to the development of GC, several studies have identified YAP1 as an oncoprotein in the stomach. In normal and proliferating normal gastric epithelium, low or moderate levels of YAP1 expression are detected, whereas significantly elevated YAP1 levels are consistently observed in both primary and metastatic GC [23, 24]. YAP1 expression is strongly positively correlated with lymphatic metastasis and shortened overall survival time, indicating that YAP may be used as an independent prognostic marker for GC [25,26,27]. Moreover, knockdown of YAP markedly suppresses cell proliferation, colony formation, and metastasis in GC cell lines, both in vitro and in vivo [28,29,30]. Besides the overexpression of YAP1, dysregulation of other genes within the Hippo signaling pathway have been reported more frequently. These include NF2, MST1/2, LATS1/2, and RASSF1A, which can increase the transcriptional activity of YAP1/TAZ/TEADs in human GC. Hippo signaling is also associated with immunosuppression [21]. On one hand, the activation of YAP1 signaling in tumor cells induces immunosuppression by up-regulating various cytokines, chemokines, and other molecules associated with the recruitment of M2 macrophages, Tregs, and MDSCs and by inhibition the infiltration and activity of NK cells and effector T-cells. On the other hand, YAP1 overexpression in immune cells facilitates immunosuppressive cell differentiation and proliferation [21]. For example, YAP1 induces the differentiation of naïve T-cells to Treg cells by up-regulating TGFBR2 [31]. In stromal cells, YAP1 promotes the transcription of myofibroblast marker genes, such as α-SMA, CYR61, and CTGF to activate cancer-associated fibroblasts (CAFs) in breast cancer. Activated YAP1 then recruits CAFs, MDSCs, and Tregs, leading to the impairment of CD8+ T-cells [32]. Our data confirmed this mechanism. We found that RP11-323N12.5 not only promoted the malignancy of human GC, but also promoted immunosuppression by upregulating YAP1 activation-related cytokines. We found that once YAP1 levels decreased in GC cells, the RP11-323N12.5-induced immunosuppressive TME was totally reversed and the percentage of Treg cells and MDSCs decreased markedly.
For decades, the “on” state of the Hippo pathway has been regarded as a tumor suppressor pathway, due to its inhibition of proliferation and promotion of apoptosis [15, 16, 28, 30]. Dysregulation of the Hippo pathway is associated with a broad spectrum of cancers, mainly due to the core protein, YAP-1 [15]. YAP1 has been identified as an oncoprotein, associated with many different types of cancer [15]. Zhang et al. showed that YAP expression is markedly elevated in clinical hepatocellular carcinoma (HCC) samples and that YAP is a key driver gene in hepatitis B virus X protein (HBx)-induced hepatocarcinogenesis in a CREB-dependent manner. YAP may serve as a novel target in HBV-associated HCC therapy [33]. Moreover, there are several clinical studies showing that YAP protein is associated with the progression and prognosis of human GC. For example, Zhang et al. first reported that YAP is strongly expressed in gastric adenocarcinomas and that YAP knockdown may inhibit gastric cancer cell proliferation and metastasis [34]. Chol et al. showed that YAP and TAZ are oncogenic initiators and drivers in human GC, with MYC as the downstream effector [35].
Besides the protein-coding genes that regulate Hippo signaling, recently, a greater number of non-coding RNAs have been reported to modulate Hippo signaling. MAYA, a lncRNA, has been reported to mediate the methylation and upstream kinase activity of MST1/2 in Hippo signaling, which consequently, leads to the failure of LATS1 phosphorylation and YAP1 target gene activation in bone metastasis [36]. A recent study also identified a lncRNA, BCAR4, as a direct transcriptional target of YAP, involved in YAP-dependent glycolysis. Mechanistically, the long-term activation of YAP up-regulates the transcription of BCAR4, which, in turn, promotes the transcription of two glycolysis-related enzymes, HK2 and PFKFB3, through Hedgehog effector GLI2/p300 complex-mediated histone acetylation, marked by H3K27ac [37]. Similar to the lncRNAs, MAYA and BCAR4, RP11-323N12.5 is also a Hippo signaling-associated lncRNA. However, RP11-323N12.5 functions differently, in that it is an enhancer of YAP1 transcription by binding to c-MYC, which is a transcriptional factor that regulates YAP1 [38].
Several studies have shown that lncRNAs can act as enhancers for certain genes by binding to DNA or protein. The lncRNA, MAGI2-AS3, prevents the development of HCC by recruiting KDM1A and promoting H3K4me2 demethylation of the RACGAP1 gene promoter [39]. Chen et al. identified an exosome-derived lncRNA, termed lymph node metastasis-associated transcript 2 (LNMAT2), which stimulates human lymphatic endothelial cell (HLEC) tube formation and migration in vitro and enhances tumor lymphangiogenesis and LN metastasis in vivo. LNMAT2 was shown to be present at high levels in bladder cancer-derived exosomes by directly interacting with heterogeneous nuclear ribonucleoprotein A2B1 (hnRNPA2B1). Subsequently, exosomal LNMAT2 is internalized by HLECs and epigenetically up-regulates prospero homeobox 1 (PROX1) expression by the recruitment of hnRNPA2B1 and by increasing H3K4 trimethylation levels in the PROX1 promoter, ultimately resulting in lymphangiogenesis and lymphatic metastasis [40].
Conclusion
In the present study, we identified the lncRNA, RP11-323N12.5, as the most significantly up-regulated lncRNA in human GC, based on data from the TCGA database. Mechanistically, RP11-323N12.5 promoted YAP1 transcription by binding to c-MYC in its promoter region. RP11-323N12.5 also promoted malignancy and immunosuppression in a YAP1 activation-dependent manner. Moreover, RP11-323N12.5 expression can be increased by YAP1/TAZ/TEADs. Therefore, RP11-323N12.5 is a novel lncRNA contributing to the increased YAP1 expression seen in human GC.
Abbreviations
- GC:
-
Gastric cancer
- lncRNA:
-
Long non-coding RNA
- TILs:
-
Tumor infiltrating leukocytes
- TCGA:
-
The Cancer Genome Atlas Program
- GTEx:
-
The Genotype-Tissue Expression
- STAD:
-
Stomach adenocarcinoma
- LPA:
-
Lysophosphatidic acid
- ISH:
-
In situ hybridization
- ChIRP:
-
Chromatin isolation by RNA purification assay
References
Lee H, Zhang Z, Krause HM. Long noncoding RNAs and repetitive elements: junk or intimate evolutionary partners? Trends Genet. 2019;35(12):892–902.
Wang J, et al. ncRNA-Encoded peptides or proteins and cancer. Mol Ther. 2019;27(10):1718–25.
Wei L, et al. The emerging role of microRNAs and long noncoding RNAs in drug resistance of hepatocellular carcinoma. Mol Cancer. 2019;18(1):147.
Bermudez M, et al. LncRNAs as regulators of autophagy and drug resistance in colorectal cancer. Front Oncol. 2019;9:1008.
Hua JT, Chen S, He HH. Landscape of noncoding RNA in prostate cancer. Trends Genet. 2019;35(11):840–51.
Fattahi S, et al. LncRNAs as potential diagnostic and prognostic biomarkers in gastric cancer: a novel approach to personalized medicine. J Cell Physiol. 2019;235(4):3189–206.
Vafadar A, et al. Long non-coding RNAs: epigenetic regulators in cancer. Curr Pharm Des. 2019;25(33):3563–77.
Zhang L, et al. Long non-coding RNAs in ocular diseases: new and potential therapeutic targets. FEBS J. 2019;286(12):2261–72.
Botti G, et al. LncRNA HOTAIR in tumor microenvironment: what role? Int J Mol Sci. 2019;20(9):2279.
Feng W, et al. ncRNAs associated with drug resistance and the therapy of digestive system neoplasms. J Cell Physiol. 2019;234(11):19143–57.
Dong P, et al. Exploring lncRNA-mediated regulatory networks in endometrial cancer cells and the tumor microenvironment: advances and challenges. Cancers. 2019;11(2):234.
Lin YH, et al. Long non-coding RNAs as mediators of tumor microenvironment and liver cancer cell communication. Int J Mol Sci. 2018;19(12):3742.
Mia MM, Singh MK. The hippo signaling pathway in cardiac development and diseases. Front Cell Dev Biol. 2019;7:211.
Zhang S, Zhou D. Role of the transcriptional coactivators YAP/TAZ in liver cancer. Curr Opin Cell Biol. 2019;61:64–71.
Zheng Y, Pan D. The hippo signaling pathway in development and disease. Dev Cell. 2019;50(3):264–82.
Pan Z, et al. The emerging role of YAP/TAZ in tumor immunity. Mol Cancer Res. 2019;17(9):1777–866.
Lin X, et al. Super-enhancer-associated LncRNA UCA1 interacts directly with AMOT to activate YAP target genes in epithelial ovarian cancer. iScience. 2019;17:242–55.
Zhu Y, et al. The MRVI1-AS1/ATF3 signaling loop sensitizes nasopharyngeal cancer cells to paclitaxel by regulating the Hippo-TAZ pathway. Oncogene. 2019;38(32):6065–81.
Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102.
Jiang R, et al. Interleukin-22 promotes human hepatocellular carcinoma by activation of STAT3. Hepatology. 2011;54(3):900–9.
Shibata M, Ham K, Hoque MO. A time for YAP1: tumorigenesis, immunosuppression and targeted therapy. Int J Cancer. 2018;143(9):2133–44.
Ni X, et al. YAP is essential for treg-mediated suppression of antitumor immunity. Cancer Discov. 2018;8(8):1026–43.
Kim SH, et al. Activating hippo pathway via rassf1 by ursolic acid suppresses the tumorigenesis of gastric cancer. Int J Mol Sci. 2019;20(19):4709.
Kim E, et al. High yes-associated protein 1 with concomitant negative LATS1/2 expression is associated with poor prognosis of advanced gastric cancer. Pathology. 2019;51(3):261–7.
Nambara S, et al. Antitumor effects of the antiparasitic agent ivermectin via inhibition of yes-associated protein 1 expression in gastric cancer. Oncotarget. 2017;8(64):107666–77.
Han XY, et al. Expression of VGLL4 and YAP protein in gastric carcinoma tissues and tumor prognosis. Minerva Med. 2018;109(6):429–35.
Lee KW, et al. Development and validation of a six-gene recurrence risk score assay for gastric cancer. Clin Cancer Res. 2016;22(24):6228–355.
Sun D, et al. YAP1 enhances cell proliferation, migration, and invasion of gastric cancer in vitro and in vivo. Oncotarget. 2016;7(49):81062–76.
Li P, et al. Elevated expression of Nodal and YAP1 is associated with poor prognosis of gastric adenocarcinoma. J Cancer Res Clin Oncol. 2016;142(8):1765–73.
Kang W, et al. Yes-associated protein 1 exhibits oncogenic property in gastric cancer and its nuclear accumulation associates with poor prognosis. Clin Cancer Res. 2011;17(8):2130–9.
Fan Y, et al. YAP-1 promotes tregs differentiation in hepatocellular carcinoma by enhancing TGFBR2 transcription. Cell Physiol Biochem. 2017;41(3):1189–98.
Zhang K, et al. Mechanical signals regulate and activate SNAIL1 protein to control the fibrogenic response of cancer-associated fibroblasts. J Cell Sci. 2016;129(10):1989–2002.
Zhang T, et al. Hepatitis B virus X protein modulates oncogene yes-associated protein by CREB to promote growth of hepatoma cells. Hepatology. 2012;56(6):2051–9.
Zhang J, et al. Expression of yes-associated protein in gastric adenocarcinoma and inhibitory effects of its knockdown on gastric cancer cell proliferation and metastasis. Int J Immunopathol Pharmacol. 2012;25(3):583–90.
Choi W, et al. YAP/TAZ initiates gastric tumorigenesis via upregulation of MYC. Cancer Res. 2018;78(12):3306–20.
Li C, et al. A ROR1-HER3-lncRNA signalling axis modulates the hippo-YAP pathway to regulate bone metastasis. Nat Cell Biol. 2017;19(2):106–19.
Zheng X, et al. LncRNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J. 2017;36(22):3325–35.
Mo X, et al. AKT1, LKB1, and YAP1 revealed as MYC interactors with NanoLuc-based protein-fragment complementation assay. Mol Pharmacol. 2017;91(4):339–47.
Pu J, et al. lncRNA MAGI2-AS3 prevents the development of HCC via recruiting KDM1A and promoting H3K4me2 demethylation of the RACGAP1 promoter. Mol Ther Nucleic Acids. 2019;18:351–62.
Chen C, et al. Exosomal long noncoding RNA LNMAT2 promotes lymphatic metastasis in bladder cancer. J Clin Invest. 2020;130(1):404–21.
Funding
This work was supported by grants from the National Natural Science Foundation (Grant numbers: 81772262 to Y.Y., 81572370, 81772569 and 81972266 to R.J.).
Author information
Authors and Affiliations
Contributions
Conception and design: JRQ and YYL. Collection and assembly of data: JJW, FH, SX, and YXS data analysis and interpretation: JJW and FH. Contribution of reagents, materials, and analysis tools wrote the paper: JJW, FH and QHZ. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared no competing interests.
Ethical approval
Animal experiments were approved and performed in accordance with the institutional guidelines for animal care of animal ethics committee of the First People’s Hospital of Kunshan (KSL-2017017). All institutional and national guidelines for the care and use of laboratory animals were followed.
Human and animal rights
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions.
Informed consent
Informed consent to be included in the study, or the equivalent, was obtained from all patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, J., Huang, F., Shi, Y. et al. RP11-323N12.5 promotes the malignancy and immunosuppression of human gastric cancer by increasing YAP1 transcription. Gastric Cancer 24, 85–102 (2021). https://doi.org/10.1007/s10120-020-01099-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-020-01099-9