Abstract
Introduction
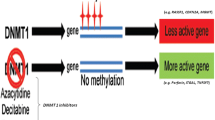
Systemic lupus erythematosus (SLE) is a heterogeneous and chronic autoimmune disease. Aberrant DNA methylation occurs during various processes of SLE development regulating the mRNA expression of interrelated genes. This study aims to screen potential DNA methylation markers for SLE.
Methods
Gene expression and methylation datasets were downloaded from the Gene Expression Omnibus (GEO) database. Differentially expressed genes (DEGs) between SLE patients and healthy controls were screened using the limma R package, and differentially methylated positions (DMPs) and regions (DMRs) were identified using dmpfinder and bumphunter (minfi). Additionally, the DNA methylation markers to distinguish SLE patients from healthy controls were explored through receiver operating characteristic (ROC) curves and logistic regression analyses. Finally, we validated the results of the bioinformatic analysis by pyrosequencing.
Results
In total, 91 DEGs, 90,092 DMPs, 15 DMRs, and 13 DMR-associated genes were identified. Through the integrative analysis of DEG- and DMR-associated genes, we identified five type I interferon (IFN)-related genes as key epigenetic-driven genes in SLE. GO enrichment analysis showed that the five SLE-associated epigenetic-driven genes were mainly enriched in the type I IFN signaling pathway involved in immune response and defense response to virus. Moreover, we identified two SLE-specific DNA methylation markers, three SLE without lupus nephritis (SLE-LN−)-specific DNA methylation markers, and two SLE with lupus nephritis (SLE-LN+)-specific DNA methylation markers by stepwise logistic regression.
Conclusions
Overall, our study demonstrates potential DNA methylation markers of SLE, SLE-LN−, and SLE-LN+, which may help the diagnosis, boost the development of new epigenetic therapy, and contribute to individualized treatment.
Key Points • This study identified five type I IFN-related genes as key epigenetic-driven genes in SLE, which support the importance of the type I IFN pathway in the pathogenesis of SLE • We identified novel DNA methylation biomarkers in SLE, SLE-LN−, and SLE-LN+ by a comprehensive analysis of bioinformatics methods and executed experimental validation, and binary logistic regression analysis showed that they have excellent potential • These results may provide new insights into the biological mechanisms of SLE, and identify reliable biomarkers for SLE, SLE-LN−, and SLE-LN+, which may contribute to diagnosis and individualized treatment |






Similar content being viewed by others
Data availability
The data that support the findings of this study are openly available in GEO (https://www.ncbi.nlm.nih.gov/geo/).
Abbreviations
- SLE:
-
Systemic lupus erythematosus
- PBMCs:
-
Peripheral blood mononuclear cells
- GEO:
-
Gene Expression Omnibus
- DEGs:
-
Differentially expressed genes
- DMPs:
-
Differentially methylated positions
- DMRs:
-
Differentially methylated regions
- ROC:
-
Receiver operating characteristic
- IFN:
-
Interferon
- LN:
-
Lupus nephritis
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- PPI:
-
Protein–protein interaction network
- AUC:
-
Area under the curve
References
Gatto M, Zen M, Ghirardello A, Bettio S, Bassi N, Iaccarino L et al (2013) Emerging and critical issues in the pathogenesis of lupus. Autoimmun Rev 12(4):523–536. https://doi.org/10.1016/j.autrev.2012.09.003
AlSaleem A, AlE’Ed A, AlSaghier A, Al-Mayouf SM (2015) Vitamin D status in children with systemic lupus erythematosus and its association with clinical and laboratory parameters. Clin Rheumatol 34(1):81–84. https://doi.org/10.1007/s10067-014-2811-z
Jiang J, Zhao M, Chang C, Wu H, Lu Q (2020) Type I interferons in the pathogenesis and treatment of autoimmune diseases. Clin Rev Allergy Immunol 59(2):248–272. https://doi.org/10.1007/s12016-020-08798-2
Jeffries MA, Sawalha AH (2015) Autoimmune disease in the epigenetic era: how has epigenetics changed our understanding of disease and how can we expect the field to evolve? Expert Rev Clin Immunol 11(1):45–58. https://doi.org/10.1586/1744666X.2015.994507
Gupta B, Hawkins RD (2015) Epigenomics of autoimmune diseases. Immunol Cell Biol 93(3):271–276. https://doi.org/10.1038/icb.2015.18
Mo XB, Zhang YH, Lei SF (2021) Integrative analysis identifies potential causal methylation-mRNA regulation chains for rheumatoid arthritis. Mol Immunol 131:89–96. https://doi.org/10.1016/j.molimm.2020.12.021
Hedrich CM, Mäbert K, Rauen T, Tsokos GC (2017) DNA methylation in systemic lupus erythematosus. Epigenomics-UK 9(4):505–525. https://doi.org/10.2217/epi-2016-0096
Jones PA, Issa JP, Baylin S (2016) Targeting the cancer epigenome for therapy. Nat Rev Genet 17(10):630–641. https://doi.org/10.1038/nrg.2016.93
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W et al (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Glickman ME, Rao SR, Schultz MR (2014) False discovery rate control is a recommended alternative to Bonferroni-type adjustments in health studies. J Clin Epidemiol 67(8):850–857. https://doi.org/10.1016/j.jclinepi.2014.03.012
Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD et al (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10):1363–1369. https://doi.org/10.1093/bioinformatics/btu049
Yang H, Wang K (2015) Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc 10(10):1556–1566. https://doi.org/10.1038/nprot.2015.105
Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H et al (2019) g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47(W1):W191–W198. https://doi.org/10.1093/nar/gkz369
Rahman A, Isenberg DA (2008) Systemic lupus erythematosus. N Engl J Med 358(9):929–939. https://doi.org/10.1056/NEJMra071297
Zhang B, Liu L, Zhou T, Shi X, Wu H, Xiang Z et al (2020) A simple and highly efficient method of IFI44L methylation detection for the diagnosis of systemic lupus erythematosus. Clin Immunol 221:108612. https://doi.org/10.1016/j.clim.2020.108612
Higgs BW, Liu Z, White B, Zhu W, White WI, Morehouse C et al (2011) Patients with systemic lupus erythematosus, myositis, rheumatoid arthritis and scleroderma share activation of a common type I interferon pathway. Ann Rheum Dis 70(11):2029–2036. https://doi.org/10.1136/ard.2011.150326
Mavragani CP, Crow MK (2010) Activation of the type I interferon pathway in primary Sjogren’s syndrome. J Autoimmun 35(3):225–231. https://doi.org/10.1016/j.jaut.2010.06.012
Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ et al (2003) Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 100(5):2610–2615. https://doi.org/10.1073/pnas.0337679100
Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J et al (2003) Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med 197(6):711–723. https://doi.org/10.1084/jem.20021553
Ding X, Ren Y, He X (2021) IFN-I mediates lupus nephritis from the beginning to renal fibrosis. Front Immunol 12:676082. https://doi.org/10.3389/fimmu.2021.676082
Kong J, Li L, Zhimin L, Yan J, Ji D, Chen Y et al (2019) Potential protein biomarkers for systemic lupus erythematosus determined by bioinformatics analysis. Comput Biol Chem 83:107135. https://doi.org/10.1016/j.compbiolchem.2019.107135
Rönnblom L, Alm GV, Eloranta ML (2011) The type I interferon system in the development of lupus. Semin Immunol 23(2):113–121. https://doi.org/10.1016/j.smim.2011.01.009
Sim TM, Ong SJ, Mak A, Tay SH (2022) Type I interferons in systemic lupus erythematosus: a journey from bench to bedside. Int J Mol Sci 23(5). https://doi.org/10.3390/ijms23052505.
Hua J, Kirou K, Lee C, Crow MK (2006) Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum 54(6):1906–1916. https://doi.org/10.1002/art.21890
Zhao M, Zhou Y, Zhu B, Wan M, Jiang T, Tan Q et al (2016) IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann Rheum Dis 75(11):1998–2006. https://doi.org/10.1136/annrheumdis-2015-208410
Nzeusseu TA, Galant C, Theate I, Maudoux AL, Lories RJ, Houssiau FA et al (2007) Identification of distinct gene expression profiles in the synovium of patients with systemic lupus erythematosus. Arthritis Rheum 56(5):1579–1588. https://doi.org/10.1002/art.22578
Diamond MS (2014) IFIT1: a dual sensor and effector molecule that detects non-2’-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev 25(5):543–550. https://doi.org/10.1016/j.cytogfr.2014.05.002
Ye S, Pang H, Gu YY, Hua J, Chen XG, Bao CD et al (2003) Protein interaction for an interferon-inducible systemic lupus associated gene, IFIT1. Rheumatology (Oxford) 42(10):1155–1163. https://doi.org/10.1093/rheumatology/keg315
Haller O, Kochs G (2011) Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. J Interferon Cytokine Res 31(1):79–87. https://doi.org/10.1089/jir.2010.0076
Feng X, Wu H, Grossman JM, Hanvivadhanakul P, FitzGerald JD, Park GS et al (2006) Association of increased interferon-inducible gene expression with disease activity and lupus nephritis in patients with systemic lupus erythematosus. Arthritis Rheum 54(9):2951–2962. https://doi.org/10.1002/art.22044
Liu X, Li H, Zhong B, Blonska M, Gorjestani S, Yan M et al (2013) USP18 inhibits NF-κB and NFAT activation during Th17 differentiation by deubiquitinating the TAK1-TAB1 complex. J Exp Med 210(8):1575–1590. https://doi.org/10.1084/jem.20122327
Fan H, Zhao G, Ren D, Liu F, Dong G, Hou Y (2017) Gender differences of B cell signature related to estrogen-induced IFI44L/BAFF in systemic lupus erythematosus. Immunol Lett 181:71–78. https://doi.org/10.1016/j.imlet.2016.12.002
Solomon DH, Kavanaugh AJ, Schur PH (2002) Evidence-based guidelines for the use of immunologic tests: antinuclear antibody testing. Arthritis Rheum 47(4):434–444. https://doi.org/10.1002/art.10561
Flechsig A, Rose T, Barkhudarova F, Strauss R, Klotsche J, Dähnrich C et al (2017) What is the clinical significance of anti-Sm antibodies in systemic lupus erythematosus? A comparison with anti-dsDNA antibodies and C3. Clin Exp Rheumatol 35(4):598–606
Morales E, Galindo M, Trujillo H, Praga M (2021) Update on lupus nephritis: looking for a new vision. Nephron 145(1):1–13. https://doi.org/10.1159/000511268
Davidson A, Aranow C, Mackay M (2019) Lupus nephritis: challenges and progress. Curr Opin Rheumatol 31(6):682–688. https://doi.org/10.1097/BOR.0000000000000642
Xu W, Xu M, Wang L, Zhou W, Xiang R, Shi Y et al (2019) Integrative analysis of DNA methylation and gene expression identified cervical cancer-specific diagnostic biomarkers. Signal Transduct Target Ther 4:55. https://doi.org/10.1038/s41392-019-0081-6
Zhao X, Zhang L, Wang J, Zhang M, Song Z, Ni B et al (2021) Identification of key biomarkers and immune infiltration in systemic lupus erythematosus by integrated bioinformatics analysis. J Transl Med 19(1):35. https://doi.org/10.1186/s12967-020-02698-x
Song G, Zhu X, Xuan Z, Zhao L, Dong H, Chen J et al (2021) Hypermethylation of GNA14 and its tumor-suppressive role in hepatitis B virus-related hepatocellular carcinoma. Theranostics 11(5):2318–2333. https://doi.org/10.7150/thno.48739
Coit P, Yalavarthi S, Ognenovski M, Zhao W, Hasni S, Wren JD et al (2015) Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J Autoimmun 58:59–66. https://doi.org/10.1016/j.jaut.2015.01.004
Xie S, Zeng Q, Ouyang S, Liang Y, Xiao C (2021) Bioinformatics analysis of epigenetic and SNP-related molecular markers in systemic lupus erythematosus. Am J Transl Res 13(6):6312–6329
Karimifar M, Pakzad B, Karimzadeh H, Mousavi M, Kazemi M, Salehi A et al (2019) Interferon-induced protein 44-like gene promoter is differentially methylated in peripheral blood mononuclear cells of systemic lupus erythematosus patients. J Res Med Sci 24:99. https://doi.org/10.4103/jrms.JRMS_83_19
Zhu H, Mi W, Luo H, Chen T, Liu S, Raman I et al (2016) Whole-genome transcription and DNA methylation analysis of peripheral blood mononuclear cells identified aberrant gene regulation pathways in systemic lupus erythematosus. Arthritis Res Ther 18:162. https://doi.org/10.1186/s13075-016-1050-x
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding
This work was supported by the National Natural Science Foundation of China [grant number 82073440] and the Medical Science and Technology Research Fund project of Guangdong Province [grant number A2019180].
Author information
Authors and Affiliations
Contributions
WJZ, ZWZ, HFZ, and KL participated in the conception and design, KL and XX provided the administrative support, HFZ, XDZ, GXL, and ZWZ participated in the provision of study materials or patients, WJZ participated in the collection and assembly of data, and data analysis and interpretation. All authors participated in manuscript writing and final approval of manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Nanfang Hospital, Southern Medical University (Guangzhou, Guangdong, China).
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to publication
All authors consent to the publication of this work.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wenjing Zhang and Guixin Liang have contributed equally to this work and share the first authorship.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, W., Liang, G., Zhou, H. et al. Identification of potential biomarkers for systemic lupus erythematosus by integrated analysis of gene expression and methylation data. Clin Rheumatol 42, 1423–1433 (2023). https://doi.org/10.1007/s10067-022-06495-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-022-06495-3