Abstract
Introduction/objectives
Persistent hyperuricemia is a key factor in gout; however, only 13.5% of hyperuricemic individuals manifest the disease. The gut microbiota could be one of the many factors underlying this phenomenon. We aimed to assess the difference in taxonomic and predicted functional profiles of the gut microbiota between asymptomatic hyperuricemia (AH) individuals and gout patients.
Methods
The V3–V4 region of the 16S rRNA gene of the gut microbiota of AH individuals, gout patients, and controls was sequenced. Bioinformatic analyses were carried out with QIIME2 and phyloseq to determine the difference in the relative abundance of bacterial genera among the study groups. Tax4fun2 was used to predict the functional profile of the gut microbiota.
Results
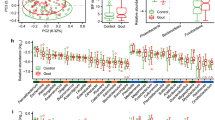
AH individuals presented a higher abundance of butyrate- and propionate-producing bacteria than gout patients; however, the latter had more bacteria capable of producing acetate. The abundance of Prevotella genus bacteria was not significantly different between the patients but was higher than that in controls. This result was corroborated by the functional profile, in which AH individuals had less pyruvate oxidase abundance than gout patients and less abundance of an enzyme that regulates glutamate synthetase activation than controls.
Conclusion
We observed a distinctive taxonomic profile in AH individuals characterized by a higher abundance of short-chain fatty acid-producing bacteria in comparison to those observed in gout patients. Furthermore, we provide scientific evidence that indicates that the gut microbiota of AH individuals could provide anti-inflammatory mediators, which prevent the appearance of gout flares.
Key Points • AH and gout patients both have a higher abundance of Prevotella genus bacteria than controls. • AH individuals’ gut microbiota had more butyrate- and propionate-producing bacteria than gout patients. • The gut microbiome of AH individuals provides anti-inflammatory mediators that could prevent gout flares. |





Similar content being viewed by others
Data availability
The data supporting the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
Lin KC, Lin HY, Chou P (2000) The interaction between uric acid level and other risk factors on the development of gout among asymptomatic hyperuricemic men in a prospective study. J Rheumatol 27:1501–1505
Johnson RJ (2015) Why focus on uric acid? Curr Med Res Opin 31(Suppl 2):3–7. https://doi.org/10.1185/03007995.2015.1087979
Shao Y, Shao H, Sawhney MS, Shi L (2019) Serum uric acid as a risk factor of all-cause mortality and cardiovascular events among type 2 diabetes population: Meta-analysis of correlational evidence. J Diabetes Complicat 33:107409. https://doi.org/10.1016/j.jdiacomp.2019.07.006
Jin M, Yang F, Yang I et al (2012) Uric acid, hyperuricemia and vascular diseases. Front Biosci 17:656–669. https://doi.org/10.2741/3950
Ichida K, Matsuo H, Takada T et al (2012) Decreased extra-renal urate excretion is a common cause of hyperuricemia. Nat Commun 3:764. https://doi.org/10.1038/ncomms1756
Xu X, Li C, Zhou P, Jiang T (n.d.) Uric acid transporters hiding in the intestine. Pharm Biol 54:3151–3155.
Schippa S, Conte M (2014) Dysbiotic events in gut microbiota: impact on human health. Nutrients 6:5786–5805. https://doi.org/10.3390/nu6125786
Duca F, a, Lam TKT, (2014) Gut microbiota, nutrient sensing and energy balance. Diabetes Obes Metab 16(Suppl 1):68–76. https://doi.org/10.1111/dom.12340
Blaak EE, Canfora EE, Theis S et al (2020) Short chain fatty acids in human gut and metabolic health. Benef Microbes 11:411–455. https://doi.org/10.3920/BM2020.0057
Cleophas MCP, Crişan TO, Lemmers H et al (2016) Suppression of monosodium urate crystal-induced cytokine production by butyrate is mediated by the inhibition of class I histone deacetylases. Ann Rheum Dis 75:593–600. https://doi.org/10.1136/ANNRHEUMDIS-2014-206258
Lim MY, Rho M, Song Y-M et al (2014) Stability of gut enterotypes in Korean monozygotic twins and their association with biomarkers and diet. Sci Rep 4:7348. https://doi.org/10.1038/srep07348
Méndez-Salazar EO, Vázquez-Mellado J, Casimiro-Soriguer CS et al (2021) Taxonomic variations in the gut microbiome of gout patients with and without tophi might have a functional impact on urate metabolism. Mol Med 27:50. https://doi.org/10.1186/s10020-021-00311-5
Guo Z, Zhang J, Wang Z et al (2016) Intestinal microbiota distinguish gout patients from healthy humans. Sci Rep 6:20602. https://doi.org/10.1038/srep20602
Shao T, Shao L, Li H et al (2017) Combined Signature of the fecal microbiome and metabolome in patients with gout. Front Microbiol 8:268. https://doi.org/10.3389/fmicb.2017.00268
Sato Y, Feig DI, Stack AG et al (2019) The case for uric acid-lowering treatment in patients with hyperuricaemia and CKD. Nat Rev Nephrol. https://doi.org/10.1038/s41581-019-0174-z
Hassan W, Shrestha P, Sumida K et al (2022) Association of uric acid-lowering therapy with incident chronic kidney disease. JAMA Netw open 5:e2215878. https://doi.org/10.1001/JAMANETWORKOPEN.2022.15878
Zhao H, Lu Z, Lu Y (2022) The potential of probiotics in the amelioration of hyperuricemia. Food Funct 13:2394–2414. https://doi.org/10.1039/D1FO03206B
Kim HW, Yoon EJ, Jeong SH, Park MC (2022) Distinct gut microbiota in patients with asymptomatic hyperuricemia: a potential protector against gout development. Yonsei Med J 63:241–251. https://doi.org/10.3349/ymj.2022.63.3.241
Yang HT, Xiu WJ, Liu JK et al (2021) Gut microbiota characterization in patients with asymptomatic hyperuricemia: probiotics increased. Bioengineered 12:7263–7275. https://doi.org/10.1080/21655979.2021.1976897
Wei J, Zhang Y, Dalbeth N et al (2022) Association between gut microbiota and elevated serum urate in two independent cohorts. Arthritis Rheumatol (Hoboken, NJ) 74:682–691. https://doi.org/10.1002/ART.42009
Neogi T, Jansen TLTA, Dalbeth N et al (2015) 2015 Gout classification criteria: an American College of Rheumatology/European League Against Rheumatism collaborative initiative. Ann Rheum Dis 74:1789–1798. https://doi.org/10.1136/annrheumdis-2015-208237
Bolyen E, Rideout JR, Dillon MR et al (n.d.) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857.
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Quast C, Pruesse E, Yilmaz P et al (n.d) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–6.
McMurdie PJ, Holmes S (2013) phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217
Willis A, Bunge J (2015) Estimating diversity via frequency ratios. Biometrics 71:1042–1049. https://doi.org/10.1111/biom.12332
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mallick H, Rahnavard A, McIver LJ, et al (2021) Multivariable association discovery in population-scale meta-omics studies. bioRxiv 2021.01.20.427420. https://doi.org/10.1101/2021.01.20.427420
Tibshirani R, Bien J, Friedman J et al (2012) Strong rules for discarding predictors in lasso-type problems. J R Stat Soc Series B Stat Methodol 74:245–266. https://doi.org/10.1111/J.1467-9868.2011.01004.X
Wemheuer F, Taylor JA, Daniel R et al (2020) Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ Microbiomes 15:1–12. https://doi.org/10.1186/S40793-020-00358-7/FIGURES/5
Parks D, Beiko R (2013) STAMP: statistical analysis of metagenomic profiles. SRC:1–6.
Wang G, Huang S, Wang Y et al (2019) Bridging intestinal immunity and gut microbiota by metabolites. Cell Mol Life Sci 76:3917–3937. https://doi.org/10.1007/S00018-019-03190-6
Zeng Q, Li D, He Y et al (2019) Discrepant gut microbiota markers for the classification of obesity-related metabolic abnormalities. Sci Rep. https://doi.org/10.1038/S41598-019-49462-W
Chu Y, Sun S, Huang Y et al (2021) Metagenomic analysis revealed the potential role of gut microbiome in gout. NPJ Biofilms Microbiomes 7:66. https://doi.org/10.1038/s41522-021-00235-2
Park HK, Lee SJ (2022) Treatment of gouty arthritis is associated with restoring the gut microbiota and promoting the production of short-chain fatty acids. Arthritis Res Ther. https://doi.org/10.1186/S13075-022-02742-9
Maeda Y, Kurakawa T, Umemoto E et al (2016) Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol 68:2646–2661. https://doi.org/10.1002/art.39783
Lucke K, Miehlke S, Jacobs E, Schuppler M (2006) Prevalence of Bacteroides and Prevotella spp. in ulcerative colitis. J Med Microbiol 55:617–624. https://doi.org/10.1099/JMM.0.46198-0
Stoll ML, Pierce MK, Watkins JA et al (n.d) Akkermansia muciniphila is permissive to arthritis in the K/BxN mouse model of arthritis. Genes Immun 20:158–166.
Wang Z, Zhu H, Jiang Q, Zhu YZ (2021) The gut microbiome as non-invasive biomarkers for identifying overweight people at risk for osteoarthritis. Microb Pathog. https://doi.org/10.1016/J.MICPATH.2021.104976
Guo Y, Yu Y, Li H et al (2021) Inulin supplementation ameliorates hyperuricemia and modulates gut microbiota in Uox-knockout mice. Eur J Nutr 60:2217–2230. https://doi.org/10.1007/S00394-020-02414-X
Vieira AT, Macia L, Galvão I et al (2015) A role for gut microbiota and the metabolite-sensing receptor GPR43 in a murine model of gout. Arthritis Rheumatol 67:1646–1656. https://doi.org/10.1002/art.39107
Park J, Kim M, Kang SG et al (2015) Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol 8:80–93. https://doi.org/10.1038/MI.2014.44
Kukolj C, Pedrosa FO, De Souza GA et al (2020) Proteomic and metabolomic analysis of azospirillum brasilense ntrC mutant under high and low nitrogen conditions. J Proteome Res 19:92–105. https://doi.org/10.1021/ACS.JPROTEOME.9B00397
Méndez-Salazar EO, Martínez-Nava GA (2022) Uric acid extrarenal excretion: the gut microbiome as an evident yet understated factor in gout development. Rheumatol Int 42:403–412. https://doi.org/10.1007/S00296-021-05007-X
Shi T, Wang Y, Wang Z et al (2014) Deregulation of purine pathway in Bacillus subtilis and its use in riboflavin biosynthesis. Microb Cell Fact 13:101. https://doi.org/10.1186/S12934-014-0101-8
Acknowledgements
We are very grateful to the participants for their time and involvement in this study. We would like to thank all staff members of the blood bank of INRLGII, especially Miguel Antonio Cervera Bustamante, Cecilia de los Angeles Nieto Gómez, Mónica Adelaida Saldaña García, María del Carmen Ramírez Flores, Graciela Morales Sánchez, Cristina Gutiérrez Mendoza, Dulce Verónica Santana Velazco, and Magdalena Gutiérrez Moreno. We also want to thank Aarón Vazquez-Mellado Montes de Oca for his logistic support in gout patient enrolment.
Funding
This study was supported by the Instituto Nacional de Rehabilitación “Luis Guillermo Ibarra Ibarra” and by the Consejo Nacional de Ciencia y Tecnología (CONACYT) grant FORDECYT-PRONACES/87754/2020 and grant INF-2016–01-269675.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics statement
The present study complies with the Declaration of Helsinki, all participants signed an informed consent letter prior to their enrolment, and the protocol was approved by INRLGII and the HGM ethics committee (INR28/15; DI/18/404-A/03/004; respectively).
Disclosures
None.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Martínez-Nava, G.A., Méndez-Salazar, E.O., Vázquez-Mellado, J. et al. The impact of short-chain fatty acid–producing bacteria of the gut microbiota in hyperuricemia and gout diagnosis. Clin Rheumatol 42, 203–214 (2023). https://doi.org/10.1007/s10067-022-06392-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-022-06392-9