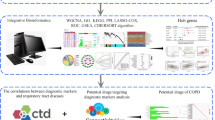
Abstract
Objective
This study aimed to identify osteoarthritis (OA) related genes based on microarray data in synovium with a more robust integrative analysis method.
Methods
Four series GSE55457, GSE12021, GSE55235, and GSE55584 (36 OA and 29 normal samples) were downloaded from the Gene Expression Omnibus database. Differentially expressed genes (DEGs) of GSE55457, GSE12021, and GSE55235 were identified using the LIMMA package. Overlapping DEGs from the intersection of the three series were detected. Simultaneously, samples in the four series were pooled to identify DEGs with integrated analysis using the Sva package.
Results
In total, 74 overlapping DEGs and 242 DEGs by integrating four series were detected. Based on them, 70 common DEGs were used to construct a protein-protein interaction (PPI) network, involving 61 nodes and 206 edges. Also, three gene modules and five hub genes, named JUN, IL6, VEGFA, MYC, and EGR1, were identified.
Conclusions
Seventy DEGs were finally identified with a more robust integrative analysis method. JUN, IL6, VEGFA, MYC, and EGR1 were identified as hub genes in the development of OA.
Key Points • 76 overlapping DEGs were detected from the intersection of DEGs in GSE55457, GSE12021, and GSE55235. • 242 DEGs were identified by integrating four series using Sva package. • 72 common DEGs were finally identified based on the overlapping DEGs and the integrated DEGs. • JUN, IL6, VEGFA, MYC, and EGR1 were identified as hub genes in the development of OA. |







Similar content being viewed by others
References
Haseeb A, Haqqi TM (2013) Immunopathogenesis of osteoarthritis. Clin Immunol (Orlando, Fla) 146(3):185–196. https://doi.org/10.1016/j.clim.2012.12.011
Heijink A, Gomoll AH, Madry H, Drobnic M, Filardo G, Espregueira-Mendes J, Van Dijk CN (2012) Biomechanical considerations in the pathogenesis of osteoarthritis of the knee. Knee Surg Sports Traumatol Arthrosc 20(3):423–435. https://doi.org/10.1007/s00167-011-1818-0
Robinson WH, Lepus CM, Wang Q, Raghu H, Mao R, Lindstrom TM, Sokolove J (2016) Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol 12(10):580–592. https://doi.org/10.1038/nrrheum.2016.136
Brandt KD, Dieppe P, Radin E (2009) Etiopathogenesis of osteoarthritis. Med Clin North Am 93(1):1–24, xv. https://doi.org/10.1016/j.mcna.2008.08.009
Jia H, Ma X, Wei Y, Tong W, Tower RJ, Chandra A, Wang L, Sun Z, Yang Z, Badar F, Zhang K, Tseng WJ, Kramer I, Kneissel M, Xia Y, Liu XS, Wang JHC, Han L, Enomoto-Iwamoto M, Qin L (2018) Loading-induced reduction in sclerostin as a mechanism of subchondral bone plate sclerosis in mouse knee joints during late-stage osteoarthritis. Arthritis Rheumatol 70(2):230–241. https://doi.org/10.1002/art.40351
Sellam J, Berenbaum F (2010) The role of synovitis in pathophysiology and clinical symptoms of osteoarthritis. Nat Rev Rheumatol 6(11):625–635. https://doi.org/10.1038/nrrheum.2010.159
He A, Ning Y, Wen Y, Cai Y, Xu K, Cai Y, Han J, Liu L, Du Y, Liang X, Li P, Fan Q, Hao J, Wang X, Guo X, Ma T, Zhang F (2018) Use of integrative epigenetic and mRNA expression analyses to identify significantly changed genes and functional pathways in osteoarthritic cartilage. Bone Joint Res 7(5):343–350. https://doi.org/10.1302/2046-3758.75.bjr-2017-0284.r1
Lin J, Wu G, Zhao Z, Huang Y, Chen J, Fu C, Ye J, Liu X (2018) Bioinformatics analysis to identify key genes and pathways influencing synovial inflammation in osteoarthritis. Mol Med Rep. https://doi.org/10.3892/mmr.2018.9575
Cai W, Li H, Zhang Y, Han G (2020) Identification of key biomarkers and immune infiltration in the synovial tissue of osteoarthritis by bioinformatics analysis. PeerJ 8:e8390. https://doi.org/10.7717/peerj.8390
Zhu N, Zhang P, Du L, Hou J, Xu B (2020) Identification of key genes and expression profiles in osteoarthritis by co-expressed network analysis. Comput Biol Chem 85:107225. https://doi.org/10.1016/j.compbiolchem.2020.107225
Li Z (2018) Integration of gene expression profile data to screen and verify hub genes involved in osteoarthritis. J Cell Biochem 2018:9482726–9487695. https://doi.org/10.1002/jcb.27118https://doi.org/10.1155/2018/9482726
Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD (2012) The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics (Oxford, England) 28(6):882–883. https://doi.org/10.1093/bioinformatics/bts034
Rahmati M, Nalesso G, Mobasheri A, Mozafari M (2017) Aging and osteoarthritis: central role of the extracellular matrix. Ageing Res Rev 40:20–30. https://doi.org/10.1016/j.arr.2017.07.004
Liu-Bryan R, Terkeltaub R (2015) Emerging regulators of the inflammatory process in osteoarthritis. Nature reviews Rheumatology 11(1):35–44. https://doi.org/10.1038/nrrheum.2014.162
Lambert C, Dubuc JE, Montell E, Verges J, Munaut C, Noel A, Henrotin Y (2014) Gene expression pattern of cells from inflamed and normal areas of osteoarthritis synovial membrane. Arthritis Rheumatol 66(4):960–968. https://doi.org/10.1002/art.38315
Scanzello CR, McKeon B, Swaim BH, DiCarlo E, Asomugha EU, Kanda V, Nair A, Lee DM, Richmond JC, Katz JN, Crow MK, Goldring SR (2011) Synovial inflammation in patients undergoing arthroscopic meniscectomy: molecular characterization and relationship to symptoms. Arthritis and rheumatism 63(2):391–400. https://doi.org/10.1002/art.30137
Favero M, Belluzzi E, Trisolino G, Goldring MB, Goldring SR, Cigolotti A, Pozzuoli A, Ruggieri P, Ramonda R, Grigolo B, Punzi L, Olivotto E (2019) Inflammatory molecules produced by meniscus and synovium in early and end-stage osteoarthritis: a coculture study. Journal of cellular physiology 234(7):11176–11187. https://doi.org/10.1002/jcp.27766
Remst DF, Blom AB, Vitters EL, Bank RA, van den Berg WB, Blaney Davidson EN, van der Kraan PM (2014) Gene expression analysis of murine and human osteoarthritis synovium reveals elevation of transforming growth factor beta-responsive genes in osteoarthritis-related fibrosis. Arthritis Rheumatol 66(3):647–656. https://doi.org/10.1002/art.38266
Latourte A, Cherifi C, Maillet J, Ea HK, Bouaziz W, Funck-Brentano T, Cohen-Solal M, Hay E, Richette P (2017) Systemic inhibition of IL-6/Stat3 signalling protects against experimental osteoarthritis. Annals of the rheumatic diseases 76(4):748–755. https://doi.org/10.1136/annrheumdis-2016-209757
Lefebvre V, Peeters-Joris C, Vaes G (1990) Modulation by interleukin 1 and tumor necrosis factor alpha of production of collagenase, tissue inhibitor of metalloproteinases and collagen types in differentiated and dedifferentiated articular chondrocytes. Biochimica et biophysica acta 1052(3):366–378
Deligne C, Casulli S, Pigenet A, Bougault C, Campillo-Gimenez L, Nourissat G, Berenbaum F, Elbim C, Houard X (2015) Differential expression of interleukin-17 and interleukin-22 in inflamed and non-inflamed synovium from osteoarthritis patients. Osteoarthritis and cartilage 23(11):1843–1852. https://doi.org/10.1016/j.joca.2014.12.007
Li Z, Zhang R, Yang X, Zhang D, Li B, Zhang D, Li Q, Xiong Y (2018) Analysis of gene expression and methylation datasets identified ADAMTS9, FKBP5, and PFKBF3 as biomarkers for osteoarthritis. Journal of cellular physiology. 234:8908–8917. https://doi.org/10.1002/jcp.27557
Fisch KM, Gamini R, Alvarez-Garcia O, Akagi R, Saito M, Muramatsu Y, Sasho T, Koziol JA, Su AI, Lotz MK (2018) Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis and cartilage 26(11):1531–1538. https://doi.org/10.1016/j.joca.2018.07.012
Rhee J, Park SH, Kim SK, Kim JH, Ha CW, Chun CH, Chun JS (2017) Inhibition of BATF/JUN transcriptional activity protects against osteoarthritic cartilage destruction. Annals of the rheumatic diseases 76(2):427–434. https://doi.org/10.1136/annrheumdis-2015-208953
Kapoor M, Martel-Pelletier J, Lajeunesse D, Pelletier JP, Fahmi H (2011) Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nature reviews Rheumatology 7(1):33–42. https://doi.org/10.1038/nrrheum.2010.196
Poree B, Kypriotou M, Chadjichristos C, Beauchef G, Renard E, Legendre F, Melin M, Gueret S, Hartmann DJ, Mallein-Gerin F, Pujol JP, Boumediene K, Galera P (2008) Interleukin-6 (IL-6) and/or soluble IL-6 receptor down-regulation of human type II collagen gene expression in articular chondrocytes requires a decrease of Sp1.Sp3 ratio and of the binding activity of both factors to the COL2A1 promoter. The Journal of biological chemistry 283(8):4850–4865. https://doi.org/10.1074/jbc.M706387200
Chen H, Tian Y (2017) MiR-15a-5p regulates viability and matrix degradation of human osteoarthritis chondrocytes via targeting VEGFA. Bioscience trends 10(6):482–488. https://doi.org/10.5582/bst.2016.01187
Macchi V, Stocco E, Stecco C, Belluzzi E, Favero M, Porzionato A, De Caro R (2018) The infrapatellar fat pad and the synovial membrane: an anatomo-functional unit. J Anat 233(2):146–154. https://doi.org/10.1111/joa.12820
Favero M, El-Hadi H, Belluzzi E, Granzotto M, Porzionato A, Sarasin G, Rambaldo A, Iacobellis C, Cigolotti A, Fontanella CG, Natali A, Ramonda R, Ruggieri P, De Caro R, Vettor R, Rossato M, Macchi V (2017) Infrapatellar fat pad features in osteoarthritis: a histopathological and molecular study. Rheumatology (Oxford) 56(10):1784–1793. https://doi.org/10.1093/rheumatology/kex287
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Fig. 1
Quality control. (a) RLE plot. (b) NUSE plot. (c) RNA degradation map. RLE relative log expression, NUSE normalized unscaled standard errors (TIF 8858 kb)
Supplementary Fig. 2
Principal component analysis. (TIF 6778 kb)
Supplementary Fig. 3
Batch correction with Sva package. (TIF 8866 kb)
Supplementary Fig. 4
The top ten significantly enriched pathways of the first two gene modules. (TIF 6366 kb)
ESM 5
(DOCX 15 kb)
ESM 6
(DOCX 18 kb)
ESM 7
(DOCX 18 kb)
ESM 8
(DOCX 18 kb)
ESM 9
(DOCX 19 kb)
ESM 10
(DOCX 15 kb)
Rights and permissions
About this article
Cite this article
Chen, H., Ni, Q., Li, B. et al. Identification of differentially expressed genes in synovial tissue of osteoarthritis based on a more robust integrative analysis method. Clin Rheumatol 40, 3745–3754 (2021). https://doi.org/10.1007/s10067-021-05649-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-021-05649-z