Abstract
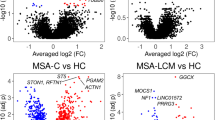
Multiple system atrophy (MSA) is a distinct member of a group of neurodegenerative diseases known as α-synucleinopathies, which are characterized by the presence of aggregated α-synuclein in the brain. MSA is unique in that the principal site for α-synuclein deposition is in the oligodendrocytes rather than neurons. The cause of MSA is unknown, and the pathogenesis of MSA is still largely speculative. Brain transcriptome perturbations during the onset and progression of MSA are mostly unknown. Using RNA sequencing, we performed a comparative transcriptome profiling analysis of the grey matter (GM) and white matter (WM) of the frontal cortex of MSA and control brains. The transcriptome sequencing revealed increased expression of the alpha and beta haemoglobin genes in MSA WM, decreased expression of the transthyretin (TTR) gene in MSA GM and numerous region-specific long intervening non-coding RNAs (lincRNAs). In contrast, we observed only moderate changes in the expression patterns of the α-synuclein (SNCA) gene, which confirmed previous observations by other research groups. Our study suggests that at the transcriptional level, MSA pathology may be related to increased iron levels in WM and perturbations of the non-coding fraction of the transcriptome.









Similar content being viewed by others
References
Wenning GK, Geser F, Krismer F, Seppi K, Duerr S, Boesch S, Kollensperger M, Goebel G, Pfeiffer KP, Barone P, Pellecchia MT, Quinn NP, Koukouni V, Fowler CJ, Schrag A, Mathias CJ, Giladi N, Gurevich T, Dupont E, Ostergaard K, Nilsson CF, Widner H, Oertel W, Eggert KM, Albanese A, del Sorbo F, Tolosa E, Cardozo A, Deuschl G, Hellriegel H, Klockgether T, Dodel R, Sampaio C, Coelho M, Djaldetti R, Melamed E, Gasser T, Kamm C, Meco G, Colosimo C, Rascol O, Meissner WG, Tison F, Poewe W, European Multiple System Atrophy Study G (2013) The natural history of multiple system atrophy: a prospective European cohort study. Lancet Neurol 12(3):264–274
Halliday GM, Holton JL, Revesz T, Dickson DW (2011) Neuropathology underlying clinical variability in patients with synucleinopathies. Acta Neuropathol 122(2):187–204. doi:10.1007/s00401-011-0852-9
Bleasel JM, Wong JH, Halliday GM, Kim WS (2014) Lipid dysfunction and pathogenesis of multiple system atrophy. Acta Neuropathol Commun 2(1):15. doi:10.1186/2051-5960-2-15
Fellner L, Jellinger KA, Wenning GK, Stefanova N (2011) Glial dysfunction in the pathogenesis of alpha-synucleinopathies: emerging concepts. Acta Neuropathol 121(6):675–693. doi:10.1007/s00401-011-0833-z
Fellner L, Stefanova N (2013) The role of glia in alpha-synucleinopathies. Mol Neurobiol 47(2):575–586. doi:10.1007/s12035-012-8340-3
Wakabayashi K, Takahashi H (2006) Cellular pathology in multiple system atrophy. Neuropathology 26(4):338–345
Wenning GK, Stefanova N, Jellinger KA, Poewe W, Schlossmacher MG (2008) Multiple system atrophy: a primary oligodendrogliopathy. Ann Neurol 64(3):239–246. doi:10.1002/ana.21465
Huang Y, Song YJ, Murphy K, Holton JL, Lashley T, Revesz T, Gai WP, Halliday GM (2008) LRRK2 and parkin immunoreactivity in multiple system atrophy inclusions. Acta Neuropathol 116(6):639–646. doi:10.1007/s00401-008-0446-3
Multiple-System Atrophy Research C (2013) Mutations in COQ2 in familial and sporadic multiple-system atrophy. N Engl J Med 369(3):233–244. doi:10.1056/NEJMoa1212115
Ozawa T, Healy DG, Abou-Sleiman PM, Ahmadi KR, Quinn N, Lees AJ, Shaw K, Wullner U, Berciano J, Moller JC, Kamm C, Burk K, Josephs KA, Barone P, Tolosa E, Goldstein DB, Wenning G, Geser F, Holton JL, Gasser T, Revesz T, Wood NW, European MSA (2006) The alpha-synuclein gene in multiple system atrophy. J Neurol Neurosurg Psychiatry 77(4):464–467. doi:10.1136/jnnp.2005.073528
Kingsbury AE, Daniel SE, Sangha H, Eisen S, Lees AJ, Foster OJ (2004) Alteration in alpha-synuclein mRNA expression in Parkinson’s disease. Mov Disord 19(2):162–170. doi:10.1002/mds.10683
Jin H, Ishikawa K, Tsunemi T, Ishiguro T, Amino T, Mizusawa H (2008) Analyses of copy number and mRNA expression level of the alpha-synuclein gene in multiple system atrophy. J Med Dent Sci 55(1):145–153
Asi YT, Simpson JE, Heath PR, Wharton SB, Lees AJ, Revesz T, Houlden H, Holton JL (2014) Alpha-synuclein mRNA expression in oligodendrocytes in MSA. Glia 62(6):964–970. doi:10.1002/glia.22653
Ozawa T, Okuizumi K, Ikeuchi T, Wakabayashi K, Takahashi H, Tsuji S (2001) Analysis of the expression level of alpha-synuclein mRNA using postmortem brain samples from pathologically confirmed cases of multiple system atrophy. Acta Neuropathol 102(2):188–190
Mills JD, Janitz M (2012) Alternative splicing of mRNA in the molecular pathology of neurodegenerative diseases. Neurobiol Aging 33(5):1012 e1011–1024. doi:10.1016/j.neurobiolaging.2011.10.030
Mills JD, Nalpathamkalam T, Jacobs HI, Janitz C, Merico D, Hu P, Janitz M (2013) RNA-Seq analysis of the parietal cortex in Alzheimer’s disease reveals alternatively spliced isoforms related to lipid metabolism. Neurosci Lett 536:90–95. doi:10.1016/j.neulet.2012.12.042
Twine NA, Janitz C, Wilkins MR, Janitz M (2013) Sequencing of hippocampal and cerebellar transcriptomes provides new insights into the complexity of gene regulation in the human brain. Neurosci Lett 541:263–268. doi:10.1016/j.neulet.2013.02.034
Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474(7351):380–384. doi:10.1038/nature10110
Langerveld AJ, Mihalko D, DeLong C, Walburn J, Ide CF (2007) Gene expression changes in postmortem tissue from the rostral pons of multiple system atrophy patients. Mov Disord 22(6):766–777. doi:10.1002/mds.21259
Wenning GK, Tison F, Seppi K, Sampaio C, Diem A, Yekhlef F, Ghorayeb I, Ory F, Galitzky M, Scaravilli T, Bozi M, Colosimo C, Gilman S, Shults CW, Quinn NP, Rascol O, Poewe W, Multiple System Atrophy Study G (2004) Development and validation of the Unified Multiple System Atrophy Rating Scale (UMSARS). Mov Disord 19(12):1391–1402. doi:10.1002/mds.20255
Morris JC (1993) The Clinical Dementia Rating (CDR): current version and scoring rules. Neurology 43(11):2412–2414
Rapaport F, Khanin R, Liang Y, Pirun M, Krek A, Zumbo P, Mason CE, Socci ND, Betel D (2013) Comprehensive evaluation of differential gene expression analysis methods for RNA-seq data. Genome Biol 14(9):R95. doi:10.1186/gb-2013-14-9-r95
Gallego Romero I, Pai AA, Tung J, Gilad Y (2014) RNA-seq: impact of RNA degradation on transcript quantification. BMC Biol 12(1):42. doi:10.1186/1741-7007-12-42
Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol. doi:10.1002/0471142727.mb1910s89, Chapter 19:Unit 19 10 11–21
Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, Miller W, Kent WJ, Nekrutenko A (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res 15(10):1451–1455. doi:10.1101/gr.4086505
Goecks J, Nekrutenko A, Taylor J (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11(8):R86. doi:10.1186/gb-2010-11-8-r86
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. doi:10.1093/bioinformatics/btp120
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. doi:10.1038/nbt.1621
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. doi:10.1038/Nprot.2008.211
Lai EC, Tam B, Rubin GM (2005) Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev 19(9):1067–1080. doi:10.1101/gad.1291905
Mills JD, Kavanagh T, Kim WS, Chen BJ, Kawahara Y, Halliday GM, Janitz M (2013) Unique transcriptome patterns of the white and grey matter corroborate structural and functional heterogeneity in the human frontal lobe. PLoS One 8(10):e78480. doi:10.1371/journal.pone.0078480
Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, Xue CH, Marinov GK, Khatun J, Williams BA, Zaleski C, Rozowsky J, Roder M, Kokocinski F, Abdelhamid RF, Alioto T, Antoshechkin I, Baer MT, Bar NS, Batut P, Bell K, Bell I, Chakrabortty S, Chen X, Chrast J, Curado J, Derrien T, Drenkow J, Dumais E, Dumais J, Duttagupta R, Falconnet E, Fastuca M, Fejes-Toth K, Ferreira P, Foissac S, Fullwood MJ, Gao H, Gonzalez D, Gordon A, Gunawardena H, Howald C, Jha S, Johnson R, Kapranov P, King B, Kingswood C, Luo OJ, Park E, Persaud K, Preall JB, Ribeca P, Risk B, Robyr D, Sammeth M, Schaffer L, See LH, Shahab A, Skancke J, Suzuki AM, Takahashi H, Tilgner H, Trout D, Walters N, Wang H, Wrobel J, Yu YB, Ruan XA, Hayashizaki Y, Harrow J, Gerstein M, Hubbard T, Reymond A, Antonarakis SE, Hannon G, Giddings MC, Ruan YJ, Wold B, Carninci P, Guigo R, Gingeras TR (2012) Landscape of transcription in human cells. Nature 489(7414):101–108. doi:10.1038/Nature11233
Li X, Buxbaum JN (2011) Transthyretin and the brain re-visited: is neuronal synthesis of transthyretin protective in Alzheimer’s disease? Mol Neurodegener 6:79. doi:10.1186/1750-1326-6-79
Schwarzman AL, Goldgaber D (1996) Interaction of transthyretin with amyloid beta-protein: binding and inhibition of amyloid formation. CIBA Found Symp 199:146–160, discussion 160–144
Santos SD, Lambertsen KL, Clausen BH, Akinc A, Alvarez R, Finsen B, Saraiva MJ (2010) CSF transthyretin neuroprotection in a mouse model of brain ischemia. J Neurochem 115(6):1434–1444. doi:10.1111/j.1471-4159.2010.07047.x
Brouillette J, Quirion R (2008) Transthyretin: a key gene involved in the maintenance of memory capacities during aging. Neurobiol Aging 29(11):1721–1732. doi:10.1016/j.neurobiolaging.2007.04.007
Celebi O, Temucin CM, Elibol B, Saka E (2014) Cognitive profiling in relation to short latency afferent inhibition of frontal cortex in multiple system atrophy. Parkinsonism Relat Disord. doi:10.1016/j.parkreldis.2014.03.012
Kao AW, Racine CA, Quitania LC, Kramer JH, Christine CW, Miller BL (2009) Cognitive and neuropsychiatric profile of the synucleinopathies: Parkinson disease, dementia with Lewy bodies, and multiple system atrophy. Alzheimer Dis Assoc Disord 23(4):365–370. doi:10.1097/WAD.0b013e3181b5065d
Kawai Y, Suenaga M, Takeda A, Ito M, Watanabe H, Tanaka F, Kato K, Fukatsu H, Naganawa S, Kato T, Ito K, Sobue G (2008) Cognitive impairments in multiple system atrophy: MSA-C vs MSA-P. Neurology 70(16 Pt 2):1390–1396. doi:10.1212/01.wnl.0000310413.04462.6a
Stankovic I, Krismer F, Jesic A, Antonini A, Benke T, Brown RG, Burn DJ, Holton JL, Kaufmann H, Kostic VS, Ling H, Meissner WG, Poewe W, Semnic M, Seppi K, Takeda A, Weintraub D, Wenning GK, On behalf of the Movement Disorders Society MSASG (2014) Cognitive impairment in multiple system atrophy: a position statement by the neuropsychology task force of the MDS multiple system atrophy (MODIMSA) study group. Mov Disord. doi:10.1002/mds.25880
Kaur D, Andersen J (2004) Does cellular iron dysregulation play a causative role in Parkinson’s disease? Ageing Res Rev 3(3):327–343. doi:10.1016/j.arr.2004.01.003
Biagioli M, Pinto M, Cesselli D, Zaninello M, Lazarevic D, Roncaglia P, Simone R, Vlachouli C, Plessy C, Bertin N, Beltrami A, Kobayashi K, Gallo V, Santoro C, Ferrer I, Rivella S, Beltrami CA, Carninci P, Raviola E, Gustincich S (2009) Unexpected expression of alpha- and beta-globin in mesencephalic dopaminergic neurons and glial cells. Proc Natl Acad Sci U S A 106(36):15454–15459. doi:10.1073/pnas.0813216106
Dexter DT, Jenner P, Schapira AH, Marsden CD (1992) Alterations in levels of iron, ferritin, and other trace metals in neurodegenerative diseases affecting the basal ganglia. The Royal Kings and Queens Parkinson’s Disease Research Group. Ann Neurol 32(Suppl):S94–S100
Abbott RD, Ross GW, Tanner CM, Andersen JK, Masaki KH, Rodriguez BL, White LR, Petrovitch H (2012) Late-life hemoglobin and the incidence of Parkinson’s disease. Neurobiol Aging 33(5):914–920. doi:10.1016/j.neurobiolaging.2010.06.023
Barbour R, Kling K, Anderson JP, Banducci K, Cole T, Diep L, Fox M, Goldstein JM, Soriano F, Seubert P, Chilcote TJ (2008) Red blood cells are the major source of alpha-synuclein in blood. Neurodegener Dis 5(2):55–59. doi:10.1159/000112832
Hoftberger R, Aboul-Enein F, Brueck W, Lucchinetti C, Rodriguez M, Schmidbauer M, Jellinger K, Lassmann H (2004) Expression of major histocompatibility complex class I molecules on the different cell types in multiple sclerosis lesions. Brain Pathol 14(1):43–50
Ishizawa K, Komori T, Arai N, Mizutani T, Hirose T (2008) Glial cytoplasmic inclusions and tissue injury in multiple system atrophy: a quantitative study in white matter (olivopontocerebellar system) and gray matter (nigrostriatal system). Neuropathology 28(3):249–257. doi:10.1111/j.1440-1789.2007.00855.x
Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci U S A 105(2):716–721. doi:10.1073/Pnas.0706729105
Faustino NA, Cooper TA (2003) Pre-mRNA splicing and human disease. Genes Dev 17(4):419–437. doi:10.1101/gad.1048803
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3(6):1101–1108
Acknowledgments
Authors wish to thank Bei Jun Chen for her contribution to GO term enrichment analysis and generation of Suppl. Fig. 1 and Caroline Janitz for her expert advice regarding Illumina sequencing. Tissues were received from the Sydney Brain Bank at Neuroscience Research Australia and the New South Wales Tissue Resource Centre at the University of Sydney which are supported by the National Health and Medical Research Council of Australia (NHMRC), University of New South Wales, Neuroscience Research Australia, Schizophrenia Research Institute and National Institute of Alcohol Abuse and Alcoholism (NIH (NIAAA) R24AA012725). This research was supported by the National Health and Medical Research Council of Australia (Project grant #1022325 to WSK and Fellowship #630434 to GMH) and Brain Foundation Australia (to MJ).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Figure 1
Schematic presentation of the RNA-Seq bioinformatics analytical pipeline. Sample 1 and 2 indicate respective biological conditions to be compared such as MSA WM, MSA GM, MSA control and WM control. Please refer to the Methods section for details. (PDF 323 kb)
Supplementary Figure 2
RT-qPCR validation of the TTR and HB genes expression pattern in GM and WM MSA samples. TTR was down-regulated 3.3-fold in MSA GM when compared to control (Con) GM (p-value<0.05). HBA1, HBA2 and HBB were up-regulated 9-, 6.3- and 10.4-fold, respectively, in MSAWM comparing to control WM (p-value<0.05). PSMB4 was used as a reference gene to normalize the expression levels of linc00263 before comparisons between tissue types were made. Expression levels were calculated using 2-ΔCt and fold-change was calculated using the 2-ΔΔCt method [50]. (PDF 50 kb)
Supplementary Table 1
Full list of genes and isoforms which show significant change in expression in MSA GM and MSA WM including GO terms enrichment analysis. (XLSX 330 kb)
Rights and permissions
About this article
Cite this article
Mills, J.D., Kim, W.S., Halliday, G.M. et al. Transcriptome analysis of grey and white matter cortical tissue in multiple system atrophy. Neurogenetics 16, 107–122 (2015). https://doi.org/10.1007/s10048-014-0430-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-014-0430-0