Abstract
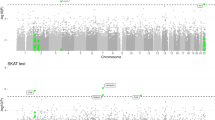
We have previously reported strong linkage on chromosome 10q in pedigrees transmitting Alzheimer's disease through the mother, overlapping with many significant linkage reports including the largest reported study. Here, we report the most comprehensive fine mapping of this region to date. In a sample of 638 late-onset Alzheimer's disease (LOAD) cases and controls including 104 maternal LOAD cases, we genotyped 3,884 single nucleotide polymorphisms (SNPs) covering 15.2 Mb. We then used imputations and publicly available data to generate an extended dataset including 4,329 SNPs for 1,209 AD cases and 839 controls in the same region. Further, we screened eight genes in this region for rare alleles in 283 individuals by nucleotide sequencing, and we tested for possible monoallelic expression as it might underlie our maternal parent of origin linkage. We excluded the possibility of multiple rare coding risk variants for these genes and monoallelic expression when we could test for it. One SNP, rs10824310 in the PRKG1 gene, showed study-wide significant association without a parent of origin effect, but the effect size estimate is not of sufficient magnitude to explain the linkage, and no association is observed in an independent genome-wide association studies (GWAS) report. Further, no causative variants were identified though sequencing. Analysis of cases with maternal disease origin pointed to a few regions of interest that included the genes PRKG1 and PCDH15 and an intergenic interval of 200 Kb. It is likely that non-transcribed rare variants or other mechanisms involving these genomic regions underlie the observed linkage and parent of origin effect. Acquiring additional support and clarifying the mechanisms of such involvement is important for AD and other complex disorder genetics research.




Similar content being viewed by others
References
Bertram L, Tanzi RE (2008) Thirty years of Alzheimer's disease genetics: the implications of systematic meta-analyses. Nat Rev Neurosci 9:768–778
Abraham R, Moskvina V, Sims R, Hollingworth P, Morgan A, Georgieva L, Dowzell K, Cichon S, Hillmer AM, O'Donovan MC et al (2008) A genome-wide association study for late-onset Alzheimer's disease using DNA pooling. BMC Med Genomics 1:44
Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Lince DH, Zismann VL, Beach TG, Leung D, Bryden L et al (2007) A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer's disease. J Clin Psychiatry 68:613–618
Li H, Wetten S, Li L, St Jean PL, Upmanyu R, Surh L, Hosford D, Barnes MR, Briley JD, Borrie M et al (2008) Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch Neurol 65:45–53
Poduslo SE, Huang R, Huang J, Smith S (2009) Genome screen of late-onset Alzheimer's extended pedigrees identifies TRPC4AP by haplotype analysis. Am J Med Genet B Neuropsychiatr Genet 150B:50–55
Carrasquillo MM, Zou F, Pankratz VS, Wilcox SL, Ma L, Walker LP, Younkin SG, Younkin CS, Younkin LH, Bisceglio GD et al (2009) Genetic variation in PCDH11X is associated with susceptibility to late-onset Alzheimer's disease. Nat Genet 41:192–198
Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, Pericak-Vance MA (2009) Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet 84:35–43
Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BM, Hooli B, Divito J, Ionita I et al (2008) Genome-wide association analysis reveals putative Alzheimer's disease susceptibility loci in addition to APOE. Am J Hum Genet 83:623–632
Grupe A, Abraham R, Li Y, Rowland C, Hollingworth P, Morgan A, Jehu L, Segurado R, Stone D, Schadt E et al (2007) Evidence for novel susceptibility genes for late-onset Alzheimer's disease from a genome-wide association study of putative functional variants. Hum Mol Genet 16:865–873
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A et al (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 41:1088–1093
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B et al (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 41:1094–1099
Potkin SG, Guffanti G, Lakatos A, Turner JA, Kruggel F, Fallon JH, Saykin AJ, Orro A, Lupoli S, Salvi E et al (2009) Hippocampal atrophy as a quantitative trait in a genome-wide association study identifying novel susceptibility genes for Alzheimer's disease. PLoS ONE 4:e6501
Reiman EM, Webster JA, Myers AJ, Hardy J, Dunckley T, Zismann VL, Joshipura KD, Pearson JV, Hu-Lince D, Huentelman MJ et al (2007) GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron 54:713–720
Bassett SS, Avramopoulos D, Fallin D (2002) Evidence for parent of origin effect in late-onset Alzheimer disease. Am J Med Genet 114:679–686
Bassett SS, Avramopoulos D, Perry RT, Wiener H, Watson B Jr, Go RC, Fallin MD (2006) Further evidence of a maternal parent-of-origin effect on chromosome 10 in late-onset Alzheimer's disease. Am J Med Genet B Neuropsychiatr Genet 141B:537–540
Blacker D, Bertram L, Saunders AJ, Moscarillo TJ, Albert MS, Wiener H, Perry RT, Collins JS, Harrell LE, Go RC et al (2003) Results of a high-resolution genome screen of 437 Alzheimer's disease families. Hum Mol Genet 12:23–32
Hamshere ML, Holmans PA, Avramopoulos D, Bassett SS, Blacker D, Bertram L, Wiener H, Rochberg N, Tanzi RE, Myers A et al (2007) Genome-wide linkage analysis of 723 affected relative pairs with late-onset Alzheimer's disease. Hum Mol Genet 16:2703–2712
Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L et al (1991) Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature 349:704–706
Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze FW, Crook R et al (2000) Susceptibility locus for Alzheimer's disease on chromosome 10. Science 290:2304–2305
Bertram L, Blacker D, Mullin K, Keeney D, Jones J, Basu S, Yhu S, McInnis MG, Go RC, Vekrellis K et al (2000) Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science 290:2302–2303
Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG (2000) Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer's disease pedigrees. Science 290:2303–2304
Wijsman EM, Daw EW, Yu CE, Payami H, Steinbart EJ, Nochlin D, Conlon EM, Bird TD, Schellenberg GD (2004) Evidence for a novel late-onset Alzheimer disease locus on chromosome 19p13.2. Am J Hum Genet 75:398–409
Bassett SS, Kusevic I, Cristinzio C, Yassa MA, Avramopoulos D, Yousem DM, Fallin MD (2005) Brain activation in offspring of AD cases corresponds to 10q linkage. Ann Neurol 58:142–146
Blacker D, Haines JL, Rodes L, Terwedow H, Go RC, Harrell LE, Perry RT, Bassett SS, Chase G, Meyers D et al (1997) ApoE-4 and age at onset of Alzheimer's disease: the NIMH genetics initiative. Neurology 48:139–147
Smith MW, Patterson N, Lautenberger JA, Truelove AL, McDonald GJ, Waliszewska A, Kessing BD, Malasky MJ, Scafe C, Le E et al (2004) A high-density admixture map for disease gene discovery in african americans. Am J Hum Genet 74:1001–1013
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Browning BL, Browning SR (2009) A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet 84:210–223
Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A 74:5463–5467
Szymanski M, Wang R, Fallin MD, Bassett SS and Avramopoulos D (2008) Neuroglobin and Alzheimer's dementia: Genetic association and gene expression changes. Neurobiol Aging (in press)
Fiscus RR (2002) Involvement of cyclic GMP and protein kinase G in the regulation of apoptosis and survival in neural cells. Neurosignals 11:175–190
Ahmed ZM, Riazuddin S, Bernstein SL, Ahmed Z, Khan S, Griffith AJ, Morell RJ, Friedman TB, Wilcox ER (2001) Mutations of the protocadherin gene PCDH15 cause Usher syndrome type 1F. Am J Hum Genet 69:25–34
Lopes AM, Ross N, Close J, Dagnall A, Amorim A, Crow TJ (2006) Inactivation status of PCDH11X: sexual dimorphisms in gene expression levels in brain. Hum Genet 119:267–275
Kimura-Yoshida C, Kitajima K, Oda-Ishii I, Tian E, Suzuki M, Yamamoto M, Suzuki T, Kobayashi M, Aizawa S, Matsuo I (2004) Characterization of the pufferfish Otx2 cis-regulators reveals evolutionarily conserved genetic mechanisms for vertebrate head specification. Development 131:57–71
Nobrega MA, Ovcharenko I, Afzal V, Rubin EM (2003) Scanning human gene deserts for long-range enhancers. Science 302:413
Uchikawa M, Takemoto T, Kamachi Y, Kondoh H (2004) Efficient identification of regulatory sequences in the chicken genome by a powerful combination of embryo electroporation and genome comparison. Mech Dev 121:1145–1158
Mahone M, Saffman EE, Lasko PF (1995) Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. Embo J 14:2043–2055
Saffman EE, Styhler S, Rother K, Li W, Richard S, Lasko P (1998) Premature translation of oskar in oocytes lacking the RNA-binding protein bicaudal-C. Mol Cell Biol 18:4855–4862
Maisonneuve C, Guilleret I, Vick P, Weber T, Andre P, Beyer T, Blum M, Constam DB (2009) Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development 136:3019–3030
Cogswell C, Price SJ, Hou X, Guay-Woodford LM, Flaherty L, Bryda EC (2003) Positional cloning of jcpk/bpk locus of the mouse. Mamm Genome 14:242–249
Creagh EM, Brumatti G, Sheridan C, Duriez PJ, Taylor RC, Cullen SP, Adrain C, Martin SJ (2009) Bicaudal is a conserved substrate for Drosophila and mammalian caspases and is essential for cell survival. PLoS ONE 4:e5055
Riemenschneider M, Mahmoodzadeh S, Eisele T, Klopp N, Schwarz S, Wagenpfeil S, Diehl J, Mueller U, Foerstl H, Illig T et al (2004) Association analysis of genes involved in cholesterol metabolism located within the linkage region on chromosome 10 and Alzheimer's disease. Neurobiol Aging 25:1305–1308
Avramopoulos D, Wang R, Valle D, Fallin MD, Bassett SS (2007) A novel gene derived from a segmental duplication shows perturbed expression in Alzheimer's disease. Neurogenetics 8:111–120
Acknowledgements
This work was supported by National Institute on Aging (NIA) grants to DA and SSB (RO1AG022099 and RO1AG021804) and an award from the Neurosciences Education and Research Foundation to DA. We thank the Harvard Brain Tissue Resource Center (MH/NS077550) and the Johns Hopkins Brain resource center for providing us with the postmortem brain tissue used for these studies. Genotyping was in part subsidized by the Broad Institute Center for Genotyping and Analysis, which is supported by grant U54 RR020278-01 from the National Center for Research Resources. Samples from the NCRAD, which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the NIA, were used in this study. We also thank Kelly Benke, Katherine Miller, and Delphine Fradin for help with data cleaning and descriptive analysis scripts. We thank contributors, including the Alzheimer's Disease Centers who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ethical standards
The experiments reported in this manuscript comply with the current laws of the country in which they were performed (USA).
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Supplementary Table 1
Power to detect association at \( \alpha = 1 \times {10^{ - {4}}} \) (DOC 31.5 kb)
Supplementary Table 2
Genotyped single nucleotide polymorphisms quality filters (DOC 28 kb)
Supplementary Table 3
Genotype counts and percentage for rs5984894 in PCDH11X. Allelic and genotype tests for association with Alzheimer's disease were not significant in the females, males, or overall (the total N of individuals with genotypes is 615). Tests for interaction with single nucleotide polymorphisms in PCDH15 (see Methods) were also not significant. (DOC 28.5 kb)
Supplementary Table 4
Top 1% of associated single nucleotide polymorphisms for maternal case–control comparison (DOC 188 kb)
Rights and permissions
About this article
Cite this article
Fallin, M.D., Szymanski, M., Wang, R. et al. Fine mapping of the chromosome 10q11-q21 linkage region in Alzheimer's disease cases and controls. Neurogenetics 11, 335–348 (2010). https://doi.org/10.1007/s10048-010-0234-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-010-0234-9