Abstract
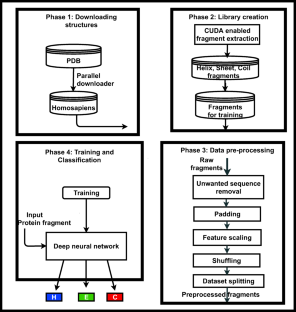
Knowledge about protein structure assignment enriches the structural and functional understanding of proteins. Accurate and reliable structure assignment data is crucial for secondary structure prediction systems. Since the 1980s, various methods based on hydrogen bond analysis and atomic coordinate geometry, followed by machine learning, have been employed in protein structure assignment. However, the assignment process becomes challenging when missing atoms are present in the protein files. Our method proposed a multi-class classifier program named DLFSA for assigning protein secondary structure elements (SSE) using convolutional neural networks (CNNs). A fast and efficient GPU-based parallel procedure extracts fragments from protein files. The model implemented in this work is trained with a subset of the protein fragments and achieves 88.1% and 82.5% train and test accuracy, respectively. The model uses only Cα coordinates for secondary structure assignments. The model has been successfully tested on a few full-length proteins also. Results from the fragment-based studies demonstrate the feasibility of applying deep learning solutions for structure assignment problems.









Similar content being viewed by others
Data availability
DLFSA is made available to the public through the Web portal—www.proteinallinfo.in. The datasets generated during and/or analyzed during the current study are not publicly available due to their large size but are available from the corresponding author on reasonable request.
References
Pauling L, Corey RB, Branson HR (1951) The structure of proteins: two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci 37(4):205–211
Reeb J, Rost B (2019) Secondary structure prediction. Encyclopedia of Bioin-formatics and Computational Biology, pp 488–496
Srinivasan R, Rose GD (1999) A physical basis for protein secondary structure. Proc Natl Acad Sci 96(25):14258–14263
Eisenberg D (2003) The discovery of the α-helix and β-sheet, the principal structural features of proteins. Proc Natl Acad Sci 100(20):11207–11210
Zhou J, Wang H, Zhao Z, Xu R, Lu Q (2018) CNNH_PSS: protein 8-class secondary structure prediction by convolutional neural network with highway. BMC Bioinform 19(4):99–109
Abbass J, Nebel JC, Mansour N, Elloumi M, Zomaya AY (2013) Ab initio protein structure prediction: methods and challenges. Biol Knowl Discov Handb. John Wiley & Sons, Inc, Hoboken, New Jersey, pp 703–724
Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181(4096):223–230
Onuchic JN, Wolynes PG (2004) Theory of protein folding. Curr Opin Struct Biol 14(1):70–75
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22(12):2577–2637
Frishman D, Argos P (1995) Knowledge-based protein secondary structure assignment. Proteins Struct Funct Bioinf 23(4):566–579
Ramachandran GT, Sasisekharan V (1968) Conformation of polypeptides and proteins. Adv Protein Chem 23:283–437
Zacharias J, Knapp EW (2014) Protein secondary structure classification revisited: processing DSSP information with PSSC. J Chem Inf Model 54(7):2166–2179
Fodje MN, Al-Karadaghi S (2002) Occurrence, conformational features and amino acid propensities for the π-helix. Protein Eng Des Sel 15(5):353–358
Nagy G, Oostenbrink C (2014) Dihedral-based segment identification and classification of biopolymers I: proteins. J Chem Inf Model 54(1):266–277
Cubellis MV, Cailliez F, Lovell SC (2005) Secondary structure assignment that accurately reflects physical and evolutionary characteristics. BMC Bioinform 6(4):1–9
Richards FM, Kundrot CE (1988) Identification of structural motifs from protein coordinate data: secondary structure and first-level supersecondary structure. Proteins Struct Funct Bioinf 3(2):71–84
Sklenar H, Etchebest C, Lavery R (1989) Describing protein structure: a general algorithm yielding complete helicoidal parameters and a unique overall axis. Proteins Struct Funct Bioinf 6(1):46–60
Hosseini SR, Sadeghi M, Pezeshk H, Eslahchi C, Habibi M (2008) PROSIGN: a method for protein secondary structure assignment based on three-dimensional coordinates of consecutive Cα atoms. Comput Biol Chem 32(6):406–411
Labesse G, Colloc'h N, Pothier J, Mornon JP (1997) P-SEA: a new efficient assignment of secondary structure from Cα trace of proteins. Bioinformatics 13(3):291–295
Majumdar I, Krishna SS, Grishin NV (2005) PALSSE: a program to delineate linear secondary structural elements from protein structures. BMC Bioinform 6(1):1–24
Taylor WR (2001) Defining linear segments in protein structure. J Mol Biol 310(5):1135–1150
Dupuis F, Sadoc JF, Mornon JP (2004) Protein secondary structure assignment through Voronoi tessellation. Proteins Struct Funct Bioinf 55(3):519–528
Park SY, Yoo MJ, Shin JM, Cho KH (2011) SABA (secondary structure assignment program based on only alpha carbons): a novel pseudo center geometrical criterion for accurate assignment of protein secondary structures. BMB Rep 44(2):118–122
Zhang W, Dunker AK, Zhou Y (2008) Assessing secondary structure assignment of protein structures by using pairwise sequence-alignment benchmarks. Proteins Struct Funct Bioinf 71(1):61–67
Cao C, Wang G, Liu A, Xu S, Wang L, Zou S (2016) A new secondary structure assignment algorithm using Cα backbone fragments. Int J Mol Sci 17(3):333
Konagurthu AS, Lesk AM, Allison L (2012) Minimum message length inference of secondary structure from protein coordinate data. Bioinformatics 28(12):i97–i105
Haghighi H, Higham J, Henchman RH (2016) Parameter-free hydrogen-bond definition to classify protein secondary structure. J Phys Chem B 120(33):8566–8570
Kumar P, Bansal M (2012) HELANAL-Plus: a web server for analysis of helix geometry in protein structures. J Biomol Struct Dyn 30(6):773–783
King SM, Johnson WC (1999) Assigning secondary structure from protein coordinate data. Proteins Struct Funct Bioinf 35(3):313–320
Carter P, Andersen CA, Rost B (2003) DSSPcont: continuous secondary structure assignments for proteins. Nucleic Acids Res 31(13):3293–3295
Konagurthu AS, Allison L, Stuckey PJ, Lesk AM (2011) Piecewise linear approximation of protein structures using the principle of minimum message length. Bioinformatics 27(13):i43–i51
Levitt M, Greer J (1977) Automatic identification of secondary structure in globular proteins. J Mol Biol 114(2):181–239
Cao C, Xu S, Wang L (2015) An algorithm for protein helix assignment using helix geometry. PLoS One 10(7):e0129674
Klose DP, Wallace BA, Janes RW (2010) 2Struc: the secondary structure server. Bioinformatics 26(20):2624–2625
Kumar P, Bansal M (2015) Identification of local variations within secondary structures of proteins. Acta Crystallogr D Biol Crystallogr 71(5):1077–1086
Habibia M, Eslahchia C, Pezeshkc H, Sadeghid M (2008) An information-theoretic approach to secondary structure assignment, Journal of Science (University of Tehran) (JSUT)
Taylor T, Rivera M, Wilson G, Vaisman II (2005) New method for protein secondary structure assignment based on a simple topological descriptor. Proteins Struct Funct Bioinf 60(3):513–524
Zhang Y, Sagui C (2015) Secondary structure assignment for conformationally irregular peptides: comparison between DSSP, STRIDE and KAKSI. J Mol Graph Model 55:72–84
Law SM, Frank AT, Brooks III CL (2014) PCASSO: a fast and efficient Cα-based method for accurately assigning protein secondary structure elements. J Comput Chem 35(24):1757–1761
Salawu EO (2016) RaFoSA: Random forests secondary structure assignment for coarse-grained and all-atom protein systems. Cogent Biol 2(1):1214061
Wang J, Cao H, Zhang JZ, Qi Y (2018) Computational protein design with deep learning neural networks. Sci Rep 8(1):1–9
Cheng J, Tegge AN, Baldi P (2008) Machine learning methods for protein structure prediction. IEEE Rev Biomed Eng 1:41–49
Zhang B, Li J, Lü Q (2018) Prediction of 8-state protein secondary structures by a novel deep learning architecture. BMC Bioinform 19(1):1–13
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444
Goh GB, Hodas NO, Vishnu A (2017) Deep learning for computational chemistry. J Comput Chem 38(16):1291–1307
O'Shea, K., & Nash, R. (2015). An introduction to convolutional neural networks. arXiv preprint arXiv:1511.08458.
Busia, A., Collins, J., & Jaitly, N. (2016). Protein secondary structure prediction using deep multi-scale convolutional neural networks and next-step conditioning. arXiv preprint arXiv:1611.01503.
Zamora-Resendiz R, Crivelli S (2019) Structural learning of proteins using graph convolutional neural networks. bioRxiv, 610444, Cold Spring Harbor Laboratory
Niepert, M., Ahmed, M., & Kutzkov, K. (2016). Learning convolutional neural networks for graphs. In International conference on machine learning (pp. 2014-2023). PMLR.
https://www.rcsb.org/structure/, accessed : 2020-09-09.
Holmes JB, Tsai J (2004) Some fundamental aspects of building protein structures from fragment libraries. Protein Sci 13(6):1636–1650
Xu D, Zhang Y (2013) Toward optimal fragment generations for ab initio protein structure assembly. Proteins Struct Funct Bioinf 81(2):229–239
de Oliveira SH, Shi J, Deane CM (2015) Building a better fragment library for de novo protein structure prediction. PLoS One 10(4):e0123998
Abbass J, Nebel JC (2015) Customised fragments libraries for protein structure prediction based on structural class annotations. BMC Bioinform 16(1):1–13
Trevizani R, Custódio FL, Dos Santos KB, Dardenne LE (2017) Critical features of fragment libraries for protein structure prediction. PLoS One 12(1):e0170131
Abbass J, Nebel JC (2020) Enhancing fragment-based protein structure prediction by customising fragment cardinality according to local secondary structure. BMC Bioinform 21:1–23
https://www.djangoproject.com/, accessed : 2020-12-12.
Acknowledgements
The authors would like to thank the Central Computing Centre, National Institute of Technology Calicut (NITC), for providing GPU servers for this work. The authors would like to acknowledge the valuable suggestions from Jinto Antony for developing the Web portal.
Code availability
The library generation source codes and the model codes are made open at https://github.com/jisnava/DLFSA/.
Funding
It is part of my (VA Jisna) PhD work at the National Institute of Technology Calicut, India. The research is funded by the Ministry of Human Resource Development, India.
Author information
Authors and Affiliations
Contributions
Jisna Vellara Antony (JVA) did the conceptualization. Hemant Yadav developed the fragment library. Prayagh Madhu did the model coding. JVA developed the Web portal and wrote the manuscript. Jayaraj Pottekkattuvalappil Balakrishnan supervised the project. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 335 kb)
Rights and permissions
About this article
Cite this article
Antony, J.V., Madhu, P., Balakrishnan, J.P. et al. Assigning secondary structure in proteins using AI. J Mol Model 27, 252 (2021). https://doi.org/10.1007/s00894-021-04825-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-021-04825-x