Abstract
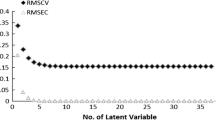
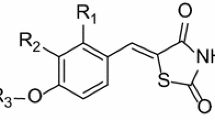
Four-dimensional quantitative structure-activity relationship (4D-QSAR) analysis was applied to a series of 52 benzothiophene analogs synthesized by Hiroshi Yamashita et al. (2011, United Sates Patent no. US8,349,840) and evaluated as dopamine D2 receptor inhibitors. The QSAR equations, generated by a combined scheme of genetic algorithms (GA) and partial least squares (PLS) regression, were evaluated by leave-one-out cross-validation, using a training and test set of 42 and ten compounds, respectively. Four different alignments were tested, and model 2, generated from Eq. 10, showed the best statistical results; it was therefore chosen to represent the data set. This study allowed a quantitative prediction of compounds potency and supported the design of the new benzothiophene.



Similar content being viewed by others
References
Silva DR, Ramalho TC, da Cunha EFF (2014) 4D-QSAR model for compounds with binding affinity towards dopamine D2 receptors. Lett Drug Des Discov 11(5):1–16
Froimowitz M, Bellott EM (1995) Structural factors that distinguish dopamine D1 and D2 agonists. J Mol Model 1:36–45
Estevinho MF, Fortunato JS (2003) Dopamine e Receptores. Rev Port Psicossomática 5:21–31
Zhang A, Neumeyer JL, Baldessarini RJ (2007) Recent progress in development of dopamine receptor subtype-selective agents: potential therapeutics for neurological and psychiatric disorders. Chem Rev 107:274–302
Seeman P, Schwarz J, Chen J, Szechtman H, Perreault M, McKnight GS, Roder JC, Quirion R, Boksa P, Srivastava LK, Yanai K, Weinshenker D, Sumiyoshi T (2006) Psychosis pathways converge via D2High dopamine receptors. Synapse 60(4):319–346
Yamashita H, Ito N, Miyamura S, Oshima K, Matsubara J, Kuroda H, Takahashi H, Shimizu S, Tanaka T (2011) Piperazine-substituted benzothiophenes for treatment of mental disorders. United Sates Patent n° US8,349,840
Damale MG, Harke SN, Khan FAK (2014) Recent advances in multidimensional QSAR (4D-6D): a critical review. Mini-Rev Med Chem 14(1):35–55
Freitas MP, Ramalho TC (2013) Employing conformational analysis in the molecular modeling of agrochemicals: insights on QSAR parameters of 2,4-D. Cienc e Agrotecnologia 37(6):485–494
Gangwal RP, Bhadauriya A, Damre M (2013) 38 mitogen-activated protein kinase inhibitors: a review on pharmacophore mapping and QSAR studies. Curr Top Med Chem 13(9):1015–1035
The Chem21 Group Inc. (1997) 4D-QSAR user’s manual. The Chem21 Group Inc., Lake forest, IL
Rogers D, Hopfinger AJ (1994) Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J Chem Inf Comput Sci 34:854–866
Romeiro NC, Albuquerque MG, Alencastro RB, Hopfinger AJ (2004) Construction of 4D-QSAR models for use in the design of novel p38-MAPK inhibitors. J Comput Aided Mol Des 19(6):385–400
da Cunha EFF, Albuquerque MG, Antunes OAC, Alencastro RB (2005) 4D-QSAR models of HOE/BAY-793 analogues as HIV-1 protease inhibitors. QSAR Comb Sci 24:240–153
Sodero ACR, Romeiro NC, da Cunha EFF, Magalhães UO, Alencastro RB, Rodrigues CR, Cabral LM, Castro HC, Albuquerque MG (2012) Application of 4D-QSAR studies to a series of raloxifene analogs and design of potential selective estrogen receptor modulators. Molecules 17:7415–7439
Hypercube, Inc. (2007) HyperChem® 7.0 software. Hypercube, Inc., Gainesville, FL
Tokarski JS, Hopfinger AJ (1997) Prediction of ligand-receptor binding thermodynamics by free energy force field (FEFF) 3D-QSAR analysis: application to a set of peptidometic renin inhibitors. J Chem Inf Comput Sci 37:792–811
Doherty DC & The Chem21Group Inc. (1997) MOLSIM Users Guide v.3.0. Doherty DC & The Chem21Group Inc., Lake Forest, IL
Weiner SJ, Kollman PA, Nguyen DT (1986) An all atom force field for simulations of proteins and nucleic acids. J Comput Chem 7:230–252
Kiralj R, Ferreira MMC (2009) Basic validation procedures for regression models in QSAR and QSPR studies: theory and application. J Braz Chem Soc 20(4):770–787
Kar S, Roy K (2011) Development and validation of a robust QSAR model for prediction of carcinogenicity of drugs. Indian J Biochem Biophys 48:111–122
Silva DG, Freitas MP, da Cunha EFF, Ramalho TC, Nunes CA (2012) Rational design of small modified peptides as ACE inhibitors. Med Chem Commun 3:1290–1293
Roy PP, Paul S, Mitra I, Roy K (2009) On two novel parameters for validation of predictive QSAR models. Molecules 14:1660–1701
Hopfinger AJ, Wang S, Tokarski JS, Jin B, Albuquerque M, Madhav PJ, Duraiswami CJ (1997) Construction of 3D-QSAR models using the 4D-QSAR analysis formalism. J Am Chem Soc 119:10509–10524
Bolton E, Wang Y, Thiessen PA, Bryant SH (2008) PubChem: Integrated platform of small molecules and biological activities Chap. 12 In: Annual Reports in Computational Chemistry, Vol. 4. American Chemical Society, Washington, DC
Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Zhou Z, Han L, Karapetyan K, Dracheva S, Shoemaker BA, Bolton E, Gindulyte A, Bryant SH (2011) PubChem’s BioAssay Database. Nucleic Acids Res 40(1):400–412
Acknowledgments
The authors are thankful to Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the financial support.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper belongs to Topical Collection Brazilian Symposium of Theoretical Chemistry (SBQT2013)
Rights and permissions
About this article
Cite this article
Caldas, G.B., Ramalho, T.C. & da Cunha, E.F.F. Application of 4D-QSAR studies to a series of benzothiophene analogs. J Mol Model 20, 2420 (2014). https://doi.org/10.1007/s00894-014-2420-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-014-2420-4