Abstract
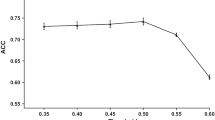
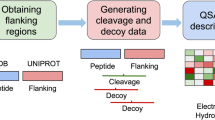
The accurate identification of cytotoxic T lymphocyte epitopes is becoming increasingly important in peptide vaccine design. The ubiquitin–proteasome system plays a key role in processing and presenting major histocompatibility complex class I restricted epitopes by degrading the antigenic protein. To enhance the specificity and efficiency of epitope prediction and identification, the recognition mode between the ubiquitin–proteasome complex and the protein antigen must be considered. Hence, a model that accurately predicts proteasomal cleavage must be established. This study proposes a new set of parameters to characterize the cleavage window and uses a backpropagation neural network algorithm to build a model that accurately predicts proteasomal cleavage. The accuracy of the prediction model, which depends on the window sizes of the cleavage, reaches 95.454 % for the N-terminus and 95.011 % for the C-terminus. The results show that the identification of proteasomal cleavage sites depends on the sequence next to it and that the prediction performance of the C-terminus is better than that of the N-terminus on average. Thus, models based on the properties of amino acids can be highly reliable and reflect the structural features of interactions between proteasomes and peptide sequences.



Similar content being viewed by others
References
Frahm N, Korber BT, Adams CM et al. (2004) Consistent cytotoxic-T-lymphocyte targeting of immuno-dominant regions in human immunodeficiency virus across multiple ethnicities. Virol 78(5):2187–2200
Takedatsu H, Shichijo S, Katagiri K et al. (2004) Identification of Peptide Vaccine Candidates Sharing among HLA-A3(+), −A11(+), −A31(+), and −A33(+). J Cancer Patients Clin Cancer Res 10(3):1112–1120
Wu YZ, Shi TD (1998) EBVD: a new approach to molecular design of vaccine. Immunologist 8(s):1487–1489
Shi TD, Wu Y (1998) Molecular design and immunological investigations on the therapeutic peptide vaccines against viral hepatitis B. Immunologist 8(s):330–334
Rosenberg SA, Yang JC, Schwartzentruber DJ et al. (1998) Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat Med 4(3):321–327
Ciechanover A (1994) The ubiquitin-proteasome proteolytic pathway. Cell 79(1):13–21
Kisselev AF, Tatos N et al. (1999) The sizes of peptides generated from protein by mammalian 26 and 20 S proteasomes. J Biol Chem 274(6):3363–3371
Coux O, Tanaka K, Goldberg AL (1996) Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem 65:801–847
Jentsch S, Schlenker S (1995) Selective protein degradation: a journey’s end within the proteasome. Cell 82(6):881–884
Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67:425–479
Groll M, Ditzel L, Lowe J et al. (1997) Structure of 20S proteasome from yeast at 2.4A resolution. Nature 386:463–471
Uebel S, Tampe R (1999) Specificity of the proteasome and the TAP transporter. Current Opinion in Immunology. J Curr Opin Immunol 11(2):203–208
Craiu A, Akopian T, Goldberg A, Rock KL (1997) Two distinct proteolytic processes in the generation of a major histocompatibility complex class I-presented peptide. Proc Natl Acad Sci 94:10850–10855
Niedermann G, King G, Butz S et al. (1996) The proteolytic fragments generated by vertebrate proteasomes: structural relationships to major histocompatibility complex class I binding peptides. Proc Natl Acad Sci 93:8572–8577
Doytchinova IA, Guan PP, Flower DR (2006) EpiJen: a server for multistep T cell epitope prediction. BMC Bioinforma 7:131–141
Pierre D, Oliver K (2005) Integrated modeling of the major events in the MHC class I antigen processing pathway. Protein Sci 14:2132–2140
Holzhutter HG, Frommel C, Kloetzel PM (1999) A theoretical approach towards the identification of cleavage-determining amino acid motifs of the 20S proteasome. J Mol Biol 286:1251–1265
Kuttler C, Nussbaum AK, Dick TP et al. (2000) An algorithm for the prediction of proteasomal cleavages. J Mol Biol 298:417–429
Nussbaum AK, Kuttler C, Hadeler KP et al. (2001) PAProC: a prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics 53:87–94
Kesmir C, Nussbaum AK, Schild H et al. (2002) Prediction of proteasome cleavage motifs by neural networks. Protein Eng 15:287–296
Altuvia Y, Margalit H (2000) Sequence signals for generation of antigenic peptides by the proteasome: implications for proteasomal cleavage mechanism. J Mol Biol 295:879–890
Goldsack DE, Chalifoux RC (1973) Contribution of the free energy of mixing of hydrophobic side chains to the stability of the tertiary structure. J Theor Biol 39:645–651
Cosic I (1994) Macromolecular bioactivity: is it resonant interaction between macromolecules?–Theory and applications. J IEEE Trans Biomed Eng 41:1101–1114
Prabhakaran M (1990) The distribution of physical, chemical and conformational properties in signal and nascent peptides. J Biochem 269:691–696
Shihab K (2006) A backpropagation neural network for computer network security. J Comput Sci 2(9):710–715
Abraham A (2005) Artificial neural networks. In: Sydenham PH, Thorn R (eds) Handbook of measuring system design. Wiley, New York, pp 901-908
Marques AJ, Palanimurugan R, Matias AC et al. (2009) Catalytic mechanism and assembly of the proteasome. Chem Rev 109(4):1509–1536
Groll M, Larionov OV, Huber R et al. (2006) Inhibitor-binding mode of homobelactosin C to proteasomes: new insights into class I MHC ligand generation. Proc Natl Acad Sci USA 103(12):4576–4579
Crooks GE, Hon G, Chandonia JM et al. (2004) WebLogo: a sequence logo generator. Genome Res 14(6):1188–1190
Saxova P, Buus S, Brunak S, Kesmir C (2003) Predicting proteasomal cleavage sites: a comparison of available methods. Int Immunol 15(7):781–787
Bhasin M, Raghava GP (2005) Pcleavage: an SVM based method for prediction of constitutive proteasome and immunoproteasome cleavage sites in antigenic sequences. Nucleic Acids Res 33(Web Server issue):W202–W207
Acknowledgments
This study was supported by grants from the National Basic Research Program of China (No. 2012CB11460), National Natural Science Foundation of China (No. 81171508, 31170747) and Natural Science Foundation Project of CQ CSTC (No. cstc2013jjb10004).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
The supplementary lists the epitope, peso-epitope and their protein.
ESM 1
(XLS 207 kb)
Rights and permissions
About this article
Cite this article
Wang, Y., Lin, Y., Shu, M. et al. Proteasomal cleavage site prediction of protein antigen using BP neural network based on a new set of amino acid descriptor. J Mol Model 19, 3045–3052 (2013). https://doi.org/10.1007/s00894-013-1827-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-013-1827-7