Abstract
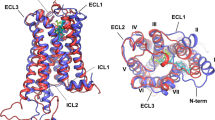
Cannabinoid and adrenergic receptors belong to the class A (similar to rhodopsin) G protein coupled receptors. Docking of agonists and antagonists to CB1 and CB2 cannabinoid receptors revealed the importance of a centrally located rotamer toggle switch and its possible participation in the mechanism of agonist/antagonist recognition. The switch is composed of two residues, F3.36 and W6.48, located on opposite transmembrane helices TM3 and TM6 in the central part of the membranous domain of cannabinoid receptors. The CB1 and CB2 receptor models were constructed based on the adenosine A2A receptor template. The two best scored conformations of each receptor were used for the docking procedure. In all poses (ligand-receptor conformations) characterized by the lowest ligand-receptor intermolecular energy and free energy of binding the ligand type matched the state of the rotamer toggle switch: antagonists maintained an inactive state of the switch, whereas agonists changed it. In case of agonists of β2AR, the (R,R) and (S,S) stereoisomers of fenoterol, the molecular dynamics simulations provided evidence of different binding modes while preserving the same average position of ligands in the binding site. The (S,S) isomer was much more labile in the binding site and only one stable hydrogen bond was created. Such dynamical binding modes may also be valid for ligands of cannabinoid receptors because of the hydrophobic nature of their ligand-receptor interactions. However, only very long molecular dynamics simulations could verify the validity of such binding modes and how they affect the process of activation.

The rotamer toggle switch in cannabinoid receptors is comprised of two residues, F3.36 and W6.48, which are located on transmembrane helices TM3 and TM6. Docking of agonists and antagonists to CB1 and CB2 cannabinoid receptors revealed the importance of this centrally located switch and its possible participation in the mechanism of agonist/antagonist sensing. The best scored poses (ligand-receptor conformations) were obtained for the ligands matching the switch state: antagonists maintained the state of the rotamer toggle switch, whereas agonists changed it






Similar content being viewed by others
References
Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX (2008) Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature 454:486–491. doi:10.1038/nature07101
Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC (2007) High-resolution crystal structure of an engineered human beta(2)-adrenergic G protein-coupled receptor. Science 318:1258–1265. doi:10.1126/science.1150577
Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK (2007) GPCR engineering yields high-resolution structural insights into beta(2)-adrenergic receptor function. Science 318:1266–1273. doi:10.1126/science.1150609
Rasmussen SGF, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, Schertler GFX, Weis WI, Kobilka BK (2007) Crystal structure of the human beta(2) adrenergic G-protein- coupled receptor. Nature 450:383–387. doi:10.1038/nature06325
Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC (2008) The 2.6 angstrom crystal structure of a human A(2A) Adenosine receptor bound to an antagonist. Science 322:1211–1217. doi:10.1126/science.1164772
Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M (2000) Crystal structure of rhodopsin: a G protein-coupled receptor. Science 289:739–745
Lei S, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, Javitch JA (2002) beta(2) adrenergic receptor activation - modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J Biol Chem 277:40989–40996. doi:10.1074/jbc.M206801200
Fritze O, Filipek S, Kuksa V, Palczewski K, Hofmann KP, Ernst OP (2003) Role of the conserved NPxxY(x)(5, 6)F motif in the rhodopsin ground state and during activation. Proc Natl Acad Sci USA 100:2290–2295. doi:10.1073/pnas.0435715100
Porter JE, Hwa J, Perez DM (1996) Activation of the alpha(1b)-adrenergic receptor is initiated by disruption of an interhelical salt bridge constraint. J Biol Chem 271:28318–28323
Kim JM, Altenbach C, Kono M, Oprian DD, Hubbell WL, Khorana HG (2004) Structural origins of constitutive activation in rhodopsin: role of the K296/E113 salt bridge. Proc Natl Acad Sci USA 101:12508–12513. doi:10.1073/pnas.0404519101
Kolinski M, Filipek S (2010) Study of a structurally similar kappa opioid receptor agonist and antagonist pair by molecular dynamics simulations. J Mol Model 16:1567–1576. doi:10.1007/s00894-010-0678-8
Kolinski M, Filipek S (2009) Studies of the activation steps concurrent to ligand binding in DOR and KOR opioid receptors based on molecular dynamics simulations. TOSBJ 3:51–63. doi:10.2174/1874199100903010051
Kolinski M, Filipek S (2008) Molecular dynamics of mu opioid receptor complexes with agonists and antagonists. TOSBJ 2:8–20. doi:10.2174/1874199100802010008
Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP, Ernst OP (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455:497–502. doi:10.1038/nature07330
Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP (2008) Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature 454:183–188. doi:10.1038/nature07063
Jozwiak K, Khalid C, Tanga MJ, Berzetei-Gurske I, Jimenez L, Kozocas JA, Woo A, Zhu W, Xiao RP, Abernethy DR, Wainer IW (2007) Comparative molecular field analysis of the binding of the stereoisomers of fenoterol and fenoterol derivatives to the beta2 adrenergic receptor. J Med Chem 50:2903–2915. doi:10.1021/jm070030d
Woo AY, Wang TB, Zeng X, Zhu W, Abernethy DR, Wainer IW, Xiao RP (2009) Stereochemistry of an agonist determines coupling preference of beta2-adrenoceptor to different G proteins in cardiomyocytes. Mol Pharmacol 75:158–165
Eswar N, Eramian D, Webb B, Shen MY, Sali A (2008) Protein structure modeling with MODELLER. Methods Mol Biol 426:145–159. doi:10.1007/978-1-60327-058-8_8
Rohl CA, Strauss CE, Misura KM, Baker D (2004) Protein structure prediction using Rosetta. Methods Enzymol 383:66–93. doi:10.1016/S0076-6879(04)83004-0
Mandell DJ, Coutsias EA, Kortemme T (2009) Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat Methods 6:551–552. doi:10.1038/nmeth0809-551
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:1–19. doi:10.1186/1471-2105-5-113
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi:10.1093/nar/gkh340
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948. doi:10.1093/bioinformatics/btm404
Gouy M, Guindon S, Gascuel O (2010) SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol 27:221–224. doi:10.1093/molbev/msp259
Ahn KH, Bertalovitz AC, Mierke DF, Kendall DA (2009) Dual role of the second extracellular loop of the Cannabinoid receptor 1: ligand binding and receptor localization. Mol Pharmacol 76:833–842. doi:10.1124/mol.109.057356
Shen MY, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524. doi:10.1110/ps.062416606
Lazaridis T (2003) Effective energy function for proteins in lipid membranes. Proteins 52:176–192
Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614
LigPrep, version 2.4: Schrödinger, LLC, New York; (2010)
Epik, version 2.1: Schrödinger, LLC, New York; (2010)
Reggio PH (2003) Pharmacophores for ligand recognition and activation/inactivation of the cannabinoid receptors. Curr Pharm Des 9:1607–1633
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi:10.1002/jcc.21256
Ballesteros JA, Weinstein H (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci 25:366–428
Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P (2003) A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem 24:1999–2012. doi:10.1002/jcc.10349
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi:10.1038/nprot.2010.5
Kolinski A (2004) Protein modeling and structure prediction with a reduced representation. Acta Biochim Pol 51:349–371
Kandt C, Ash WL, Tieleman DP (2007) Setting up and running molecular dynamics simulations of membrane proteins. Methods 41:475–488. doi:10.1016/j.ymeth.2006.08.006
Van der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718. doi:10.1002/jcc.20291
Oostenbrink C, Villa A, Mark AE, van Gunsteren WF (2004) A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 25:1656–1676. doi:10.1002/jcc.20090
Kukol A (2009) Lipid models for united-atom molecular dynamics simulations of proteins. J Chem Theory Comput 5:615–626. doi:10.1021/ct8003468
van der Spoel D, van Maaren PJ, Berendsen HJC (1998) A systematic study of water models for molecular simulation: derivation of water models optimized for use with a reaction field. J Chem Phys 108:10220–10230. doi:10.1063/1.476482
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. doi:10.1063/1.464397
Hess B, Bekker H, Berendsen HJC, Fraaije J (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Schuttelkopf AW, van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. doi:10.1107/S0907444904011679
Gouldson P, Calandra B, Legoux P, Kerneis A, Rinaldi-Carmona M, Barth F, Le Fur G, Ferrara P, Shire D (2000) Mutational analysis and molecular modelling of the antagonist SR 144528 binding site on the human cannabinoid CB(2) receptor. Eur J Pharmacol 401:17–25. doi:10.1016/S0014-2999(00)00439-8
Song ZH, Slowey CA, Hurst DP, Reggio PH (1999) The difference between the CB(1) and CB(2) cannabinoid receptors at position 5.46 is crucial for the selectivity of WIN55212-2 for CB(2). Mol Pharmacol 56:834–840
Wieland K, Zuurmond HM, Krasel C, Ijzerman AP, Lohse MJ (1996) Involvement of Asn-293 in stereospecific agonist recognition and in activation of the beta(2)-adrenergic receptor. Proc Natl Acad Sci USA 93:9276–9281
McAllister SD, Hurst DP, Barnett-Norris J, Lynch D, Reggio PH, Abood ME (2004) Structural mimicry in class A G protein-coupled receptor rotamer toggle switches - the importance of the F3.36(201)/W6.48(357) interaction in cannabinoid CB1 receptor activation. J Biol Chem 279:48024–48037. doi:10.1074/jbc.M406648200
Jeffrey GA (1997) An introduction to Hydrogen bonding. Oxford University Press, Oxford
Gonzalez A, Duran LS, Araya-Secchi R, Garate JA, Pessoa-Mahana CD, Lagos CF, Perez-Acle T (2008) Computational modeling study of functional microdomains in cannabinoid receptor type 1. Bioorg Med Chem 16:4378–4389. doi:10.1016/j.bmc.2008.02.070
McAllister SD, Rizvi G, Anavi-Goffer S, Hurst DP, Barnett-Norris J, Lynch DL, Reggio PH, Abood ME (2003) An aromatic microdomain at the cannabinoid CB1 receptor constitutes an agonist/inverse agonist binding region. J Med Chem 46:5139–5152. doi:10.1021/jm0302647
Durdagi S, Papadopoulos MG, Zoumpoulakis PG, Koukoulitsa C, Mavromoustakos T (2010) A computational study on cannabinoid receptors and potent bioactive cannabinoid ligands: homology modeling, docking, de novo drug design and molecular dynamics analysis. Mol Divers 14:257–276. doi:10.1007/s11030-009-9166-4
Shim JY (2009) Transmembrane helical domain of the Cannabinoid CB1 receptor. Biophys J 96:3251–3262. doi:10.1016/j.bpj.2008.12.3934
Shim JY (2010) Understanding functional residues of the Cannabinoid CB1 receptor for drug discovery. Curr Top Med Chem 10:779–798
Poso A, Huffman JW (2008) Targeting the cannabinoid CB2 receptor: modelling and structural determinants of CB2 selective ligands. Br J Pharmacol 153:335–346. doi:10.1038/sj.bjp.0707567
Tuccinardi T, Ferrarini PL, Manera C, Ortore G, Saccomanni G, Martinelli A (2006) Cannabinoid CB2/CB1 selectivity. Receptor modeling and automated docking analysis. J Med Chem 49:984–994. doi:10.1021/jm050875u
Seifert R, Dove S (2009) Functional selectivity of GPCR ligand stereoisomers: new pharmacological opportunities. Mol Pharmacol 75:13–18. doi:10.1124/mol.108.052944
Yao XJ, Parnot C, Deupi X, Ratnala VRP, Swaminath G, Farrens D, Kobilka B (2006) Coupling ligand structure to specific conformational switches in the beta(2)-adrenoceptor. Nat Chem Biol 2:417–422. doi:10.1038/nchembio801
Ghanouni P, Gryczynski Z, Steenhuis JJ, Lee TW, Farrens DL, Lakowicz JR, Kobilka BK (2001) Functionally different agonists induce distinct conformations in the G protein coupling domain of the beta 2 adrenergic receptor. J Biol Chem 276:24433–24436. doi:10.1074/jbc.C100162200
Swaminath G, Xiang Y, Lee TW, Steenhuis J, Parnot C, Kobilka BK (2004) Sequential binding of agonists to the beta2 adrenoceptor. Kinetic evidence for intermediate conformational states. J Biol Chem 279:686–691. doi:10.1074/jbc.M310888200
Swaminath G, Deupi X, Lee TW, Zhu W, Thian FS, Kobilka TS, Kobilka B (2005) Probing the beta2 adrenoceptor binding site with catechol reveals differences in binding and activation by agonists and partial agonists. J Biol Chem 280:22165–22171. doi:10.1074/jbc.M502352200
Romo TD, Grossfield A, Pitman MC (2010) Concerted interconversion between ionic lock substates of the beta(2) adrenergic receptor revealed by microsecond timescale molecular dynamics. Biophys J 98:76–84. doi:10.1016/j.bpj.2009.09.046
Bokoch MP, Zou Y, Rasmussen SG, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi HJ, Thian FS, Kobilka TS, Puglisi JD, Weis WI, Pardo L, Prosser RS, Mueller L, Kobilka BK (2010) Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463:108–112. doi:10.1038/nature08650
Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EYT, Velasquez J, Kuhn P, Stevens RC (2008) A specific cholesterol binding site is established by the 2.8 angstrom structure of the human beta(2)-adrenergic receptor. Structure 16:897–905
Nebane NM, Kellie B, Song ZH (2006) The effects of charge-neutralizing mutation D6.30N on the functions of CB1 and CB2 cannabinoid receptors. FEBS Lett 580:5392–5398. doi:10.1016/j.febslet.2006.09.001
Dror RO, Arlow DH, Borhani DW, Jensen MO, Piana S, Shaw DE (2009) Identification of two distinct inactive conformations of the beta(2)-adrenergic receptor reconciles structural and biochemical observations. Proc Natl Acad Sci USA 106:4689–4694. doi:10.1073/pnas.0811065106
Acknowledgments
The work was supported by the Polish Ministry of Science and Higher Education (Grant no. N N301 2038 33) and by the Foundation for Polish Science (FOCUS and TEAM programmes). Publication supported by the European Union (Innovative Economy - European Regional Development Fund) grant no. POIG.02.03.00-00-003/09. We also acknowledge the computational grants no. G07-13 and G35-6 awarded by the Interdisciplinary Centre for Mathematical and Computational Modelling in Warsaw.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
Figure S1
The alignment of human CB1 and CB2 receptor sequences with these of A2AR (a) and β1AR (b) templates. Transmembrane helices of templates are encircled (red dashed mline), the conserved residues (x.50) in each helix are in blue and cysteine residues forming disulfide bridge are in yellow. Conserved sequence motifs in cannabinoid receptors are underlined (PDF 1400 kb)
Figure S2
The representative plots of χ 1 torsion angle for residues in the rotamer toggle switch (F3.36 and W6.48) for the best scored receptor models based on the A2AR template from Modeller. (a) and (b) CB1 receptor; (c) and (d) CB2 receptor. Molecular dynamics simulations were conducted using IMM1 method (employing implicit membrane) in CHARMM program. Frames were saved every 1 ps (PDF 355 kb)
Figure S3
The RMSD plots of complexes of cannabinoid receptors during 5.5 ns MD simulations in explicit POPC membrane in YASARA program. Red line indicates plot for Cα atoms of transmembrane region; green line – plot for Cα atoms of residues from the binding site of the receptor. Frames were saved every 25 ps (PDF 310 kb)
Rights and permissions
About this article
Cite this article
Latek, D., Kolinski, M., Ghoshdastider, U. et al. Modeling of ligand binding to G protein coupled receptors: cannabinoid CB1, CB2 and adrenergic β2AR. J Mol Model 17, 2353–2366 (2011). https://doi.org/10.1007/s00894-011-0986-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-0986-7