Abstract
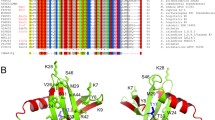
Sulfolobus synthesizes large amounts of small chromatin proteins Cren7 and Sul7d. The two proteins share overall structural similarity, but differ distinctly in the DNA-binding region between β3- and β4-strands. While Sul7d possesses a hinge of two amino acid residues, Cren7 contains a flexible seven-residue loop (loop β3–β4) in the region. Here, we report the role of loop β3–β4 in the interaction of Cren7 with duplex DNA. We show that all residues with a large side chain on the loop, i.e., Pro30, Lys31, Arg33 and Lys34, contributed significantly to the binding of Cren7 to DNA. The three basic amino acids affected the ability of Cren7 to constrain negative DNA supercoils in a residue number-dependent manner. The crystal structure of a complex between a mutant Cren7 protein (GR) with loop β3–β4 replaced by two residues (Gly and Arg) to mimic the hinge at the corresponding position in Sul7d and an 8-bp dsDNA has been determined. Structural comparison between the GR–DNA and Cren7–DNA complexes shows that GR resembles Sul7d more than Cren7 in DNA-binding size and in the effect on the width of the major groove of DNA and the pattern of DNA bending. However, GR induces smaller DNA curvature than Sul7d. Our results suggest that Cren7 and Sul7d package chromosomal DNA in a slightly different fashion, presumably permitting different chromosomal accessibility by proteins functioning in DNA transactions.





Similar content being viewed by others
References
Arnott S (1999) Oxford Handbook of Nucleic Acid Structure. Oxford University Press, Oxford
Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296:148–151
Collaborative Computational Project N (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763
Cubonova Lu, Sandman K, Hallam SJ, Delong EF, Reeve JN (2005) Histones in crenarchaea. J Bacteriol 187:5482–5485
Dame RT (2005) The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol 56:858–870
Driessen RPC et al (2013) Crenarchaeal chromatin proteins Cren7 and Sul7 compact DNA by inducing rigid bends. Nucleic Acids Res 41:196–205
Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132
Feng Y, Yao H, Wang J (2010) Crystal structure of the crenarchaeal conserved chromatin protein Cren7 and double-stranded DNA complex. Protein Sci 19:1253–1257
Gao YG et al (1998) The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA. Nat Struct Biol 5:782–786
Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15:305–308
Grant PA (2001) A tale of histone modifications. Genome Biol 2: REVIEWS0003
Grote M, Dijk J, Reinhardt R (1986) Ribosomal and DNA-Binding Proteins of the Thermoacidophilic Archaebacterium Sulfolobus Acidocaldarius. Biochim Biophys Acta 873:405–413. doi:10.1016/0167-4838(86)90090-7
Guo R, Xue H, Huang L (2003) Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo. Mol Microbiol 50:1605–1615
Guo L, Feng Y, Zhang Z, Yao H, Luo Y, Wang J, Huang L (2008) Biochemical and structural characterization of Cren7, a novel chromatin protein conserved among Crenarchaea. Nucleic Acids Res 36:1129–1137
Hansen JC, Lu X, Ross ED, Woody RW (2006) Intrinsic protein disorder, amino acid composition, and histone terminal domains. J Biol Chem 281:1853–1856
Kuhlman B, O’Neill JW, Kim DE, Zhang KY, Baker D (2001) Conversion of monomeric protein L to an obligate dimer by computational protein design. Proc Natl Acad Sci USA 98:10687–10691
Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) Procheck—a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi:10.1107/S0021889892009944
Lu X-J, Olson WK (2008) 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc 3:1213–1227
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251–260
Luijsterburg MS, White MF, van Driel R, Dame RT (2008) The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit Rev Biochem Mol Biol 43:393–418
Luo X, Schwarz-Linek U, Botting CH, Hensel R, Siebers B, White MF (2007) CC1, a novel crenarchaeal DNA binding protein. J Bacteriol 189:403–409
Mai VQ, Chen X, Hong R, Huang L (1998) Small abundant DNA binding proteins from the thermoacidophilic archaeon Sulfolobus shibatae constrain negative DNA supercoils. J Bacteriol 180:2560–2563
Marsh VL, Peak-Chew SY, Bell SD (2005) Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J Biol Chem 280:21122–21128
McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi:10.1107/S0021889807021206
Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255
Napoli A, Zivanovic Y, Bocs C, Buhler C, Rossi M, Forterre P, Ciaramella M (2002) DNA bending, compaction and negative supercoiling by the architectural protein Sso7d of Sulfolobus solfataricus. Nucleic Acids Res 30:2656–2662
Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 276:307–326. doi:10.1016/S0076-6879(97)76066-X
Reeve JN (2003) Archaeal chromatin and transcription. Mol Microbiol 48:587–598
Robinson H, Gao YG, McCrary BS, Edmondson SP, Shriver JW, Wang AH (1998) The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 392:202–205
Sandman K, Reeve JN (2005) Archaeal chromatin proteins: different structures but common function? Curr Opin Microbiol 8:656–661
Wu LJ (2004) Structure and segregation of the bacterial nucleoid. Curr Opin Genet Dev 14:126–132
Zhang Z, Gong Y, Guo L, Jiang T, Huang L (2010) Structural insights into the interaction of the crenarchaeal chromatin protein Cren7 with DNA. Mol Microbiol 76:749–759
Zhang Z, Guo L, Huang L (2012) Archaeal chromatin proteins. Sci China Life Sci 55:377–385
Acknowledgments
We are grateful to Dr. Peng Cao of the Institute of Biophysics, Chinese Academy of Sciences for her help in structure refinement. This work was supported by grant 31130003 to Li Huang and grants 30900023 and 31270124 to Zhenfeng Zhang from the National Natural Science Foundation of China.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Atomi.
The atomic coordinates and structure factors for the GR–GTGATCAC and the Cren7–GTGATCAC complexes have been deposited in the Protein Data Bank (http://www.pdb.org) under accession codes 4R55 and 4R56, respectively.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Z., Gong, Y., Chen, Y. et al. Insights into the interaction between Cren7 and DNA: the role of loop β3–β4. Extremophiles 19, 395–406 (2015). https://doi.org/10.1007/s00792-014-0725-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-014-0725-y