Abstract
Bacterial type IV pili perform important functions in such disparate biological processes as surface adhesion, cell–cell interactions, autoaggregation, conjugation, and twitching motility. Unlike bacteria, archaea use a type IV pilus related structure to drive swimming motility. While this unique flagellum is the best-studied example of an archaeal IV pilus-like structure, recent in silico, in vivo and structural analyses have revealed a highly diverse set of archaeal non-flagellar type IV pilus-like structures. Accumulating evidence suggests that these structures play important diverse roles in archaea.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Archaea are best known for their ability to thrive in extreme environments such as those having elevated temperatures, saturating salt concentrations, or extremely acidic pH. However, recent molecular studies have shown that organisms from this prokaryotic domain of life are ubiquitous (Chaban et al. 2006a). In addition to inhabiting environments such as the hot springs of Yellowstone Park and the frozen tundra of Siberia, archaea have been identified in nearly every habitat in which life of any kind has been found, including environments that range from garden soil to oral cavities and from coral reefs to the human gut (Falkowski et al. 2008; Robertson et al. 2005; Rosenberg et al. 2007).
In addition to playing important roles in essential environmental processes such as the carbon and nitrogen cycles, accumulating evidence indicates that archaea can also have significant impacts on human health. This has resulted in more intense efforts to identify the molecular mechanisms underlying key archaeal cellular processes (Conway de Macario and Macario 2008; Francis et al. 2007; Lepp et al. 2004; Pennisi 2005; Scanlan et al. 2008; Vianna et al. 2008). In that regard, recent in silico, biochemical and genetic analyses have led to a greater appreciation and understanding of the diversity and biosynthesis of archaeal type IV pilus-like structures. These pilin structures in bacteria are known to play crucial roles in fundamental cellular processes (Burrows 2005; Craig and Li 2008; Pelicic 2008).
Type IV pili, which were first identified in gram-negative bacteria, are thin filamentous protein complexes that form flexible filaments. These filaments, which traverse the outer cell membrane, can extend from the cell surface, attach to a variety of surfaces and retract, allowing them to facilitate such disparate and important functions as surface adhesion and cell–cell interactions, which mediate processes that include autoaggregation and conjugation, and twitching motility (surface gliding). Type IV pili can also attach to a pilus and are drawn close to the cell when the pilus retracts (Burrows 2005; Craig and Li 2008; Pelicic 2008).
Unlike bacterial flagella, archaeal flagella are synthesized in a manner that is similar to the synthesis of bacterial type IV pili (Ng et al. 2008; Jarrell and McBride 2008; Trachtenberg and Cohen-Krausz 2006; Albers et al. 2006). In fact, the protein subunits of archaeal flagella bear a resemblance to bacterial type IV pilins. Moreover, recent in silico analyses of more than twenty archaeal genomes have identified a large number of archaeal genes that encode putative proteins resembling type IV pilins (Szabó et al. 2007b). The expression of several archaeal pilin-like proteins has since been confirmed in vivo (Wang et al. 2008; Zolghadr et al. 2007; Fröls et al. 2007, 2008). The sequence divergence of these proteins as well as the differential expression of the operons encoding these proteins suggests they play a variety of roles in distinct biological processes.
Structure and biosynthesis of type IV pilin-like structures
Typically, a bacterial type IV pilus is composed of major pilins, the primary structural components of the filament, and several minor pilin-like proteins thought to be involved in the assembly or function of the pilus (Craig and Li 2008; Hansen and Forest 2006; Proft and Baker 2009).
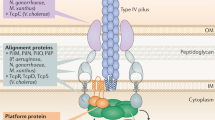
Many of the details of pilus assembly have been delineated in bacteria. Pilin precursor proteins are transported across the cytoplasmic membrane via the Sec translocation pathway (Arts et al. 2007; Francetic et al. 2007). However, while most Sec substrates are then cleaved at processing sites subsequent to the hydrophobic portion of the signal peptide by signal peptidase I or signal peptidase II, a dedicated prepilin peptidase (SPase III) cleaves pilin precursors at sites that precede the hydrophobic domain, removing only the positively charged cytoplasmic portion of the signal peptide, resulting in mature pilins having highly hydrophobic N-termini (Fig. 1) (Bardy et al. 2003; Ng et al. 2007; Pohlschroder et al. 2005). The hydrophobic nature of these N-termini is extremely important to the assembly of a pilus since extracellular interactions between these domains drives the formation of a hydrophobic central core, providing a scaffold for the assembly of the entire pilus complex (see below). A negatively charged amino acid found at position +5 within the hydrophobic domain also appears to be important for the assembly of most type IV pili. The SPase III processing site includes G/A at position −1 and a phenylalanine at position +1, which upon cleavage by SPase III is methylated by this peptidase (Peabody et al. 2003).
In addition to the prepilin peptidase, an ATPase (PilB), which provides the energy necessary for the pilus assembly, and a multispanning membrane protein (PilC), which may serve as a pilus assembly platform, are crucial to the assembly of a pilus filament (Fig. 1a). In addition, PilT, an ATPase that is an antagonist of PilB, stimulates pilus retraction. The mechanism that couples ATP hydrolysis to pilus assembly and disassembly has not yet been determined. In gram-negative bacteria, the assembly of functional type IV pili and related structures requires several additional periplasmic components, e.g. PilQ, an outer membrane pore (Fig. 1a) (Craig et al. 2006; Hansen and Forest 2006; Pelicic 2008).
Processing and maturation of archaeal class III signal peptide-containing proteins
The most extensively studied archaeal proteins containing class III signal peptides are the archaeal flagellins. Just as in bacteria, rotating flagella are responsible for archaeal swimming motility; however, in contrast to bacterial flagellar subunits, which are translocated via a specialized type III secretion apparatus, archaeal flagellin secretion and flagellar assembly resemble the processes used to translocate type IV pilins and assemble type IV pili (Jarrell and McBride 2008; Trachtenberg and Cohen-Krausz 2006). The processing of archaeal flagellins by an archaeal SPase III-like signal peptidase, FlaK, was first demonstrated in vivo in Methanococcus ssp. and later confirmed in vitro in Methanococcus maripaludis and Methanococcus voltae (Correia and Jarrell 2000; Bardy and Jarrell 2002, 2003). While heterologous expression analysis of the Sulfolobus solfataricus FlaK homolog (PibD) confirmed that it cleaves crenarchaeal flagellin precursors, attempts to generate pibD deletion mutants have failed, possibly due to the fact that it also is essential for processing other class III signal peptides (Albers et al. 2003). While G/A is conserved at −1 in the FlaK recognition site and bacterial SPase III recognition site sequences, FlaK processing sites have an uncharged amino acid at +1 and often lack the positively charged amino acid at position +5 that is found in bacterial type IV pilins. However, amino acids at positions −2, and to a certain extent −3, relative to the cleavage site appear to be crucial for recognition and processing by FlaK (Thomas et al. 2002).
FlaFind, a perl program designed to detect archaeal class III signal peptides, has been used to identify genes that encode non-flagellin archaeal pilin-like proteins (Szabó et al. 2007b). Interestingly, many FlaFind positives encode proteins containing a relatively unique SPase III signal peptidase recognition site that, reminiscent of the bacterial type IV pilus contains a +5 negative charge (−2[K/A] − 1[G/A] + [Q] + [2]X + 3[S/T/A] + 4[X] + 5[E/D]). Operons that encode these proteins often also contain a gene that encodes a novel FlaK paralog (EppA), which has four extra transmembrane domains. Heterologous analysis has revealed that M. maripaludis proteins containing the unique SPase III recognition site (EpdA and EpdC) are processed specifically by the apparently co-regulated EppA homolog. Conversely, M. maripaludis preflagellins are cleaved only by the archaeal preflagellin peptidase FlaK. Consistent with its apparent specificity, EppA homologs are found exclusively in organisms that express proteins containing the unique EppA-recognition signal (Szabó et al. 2007b). Recently, it was shown that EpdA and EpdC are indeed required for pilus biosynthesis (Wang et al. 2008). Deletion of the genes encoding these EppA substrates results in M. maripaludis cells lacking pili that can be detected on the surface of wild-type M. maripaludis that are grown under standard laboratory conditions (Ng S, unpublished results).
While the EppAs appear to specifically cleave substrates with this conserved motif, and the M. maripaludis FlaK is a flagellin-specific peptidase, the S. solfataricus FlaK homolog, PibD, exhibits broader substrate specificity. For example, it has been shown that this SPase III can process the class III signal peptides of two subunits of an S. solfataricus type IV pilus-like structure, UpsA and UpsB, that are induced by UV exposure (Fig. 1b) (Fröls et al. 2008) (see below).
Moreover, the S. solfataricus genome contains eight FlaFind positive genes that encode putative substrate-binding proteins (SBPs); of these, PibD has already been shown to cleave GlcS, AraS and TreS. Given that these putative proteins contain class III signal peptides, it has been suggested that, once processed and glycosylated, these SBPs might be assembled into a macromolecular structure, tentatively called a bindosome, that integrates into the cell membrane (Albers et al. 2003) (see below). Interestingly, these proteins are significantly larger than known pilins, containing approximately 550 amino acids as compared to the 150–200 amino acids contained in pilins. The broad substrate specificity of PibD does not appear to be specific to S. solfataricus, as several other crenarchaea and euryarchaea, which have genomes that encode a diverse set of putative proteins containing class III peptides, appear to express a single SPase III homolog (Szabó et al. 2007b).
Assembly machinery of archaeal type IV pilus-like structures
As in bacterial type IV pili assembly, in addition to a prepilin peptidase, the assembly of euryarchaeal and crenarchaeal flagella requires an ATPase homologous to PilB (FlaI) and a multispanning membrane protein (FlaJ) having some homology to PilC (Chaban et al. 2006b; Bardy et al. 2004; Jarrell and McBride 2008; Patenge et al. 2001; Thomas et al. 2002). UV-induced pilus biosynthesis in S. solfataricus requires UpsE, a PilB paralog (distinct from FlaI) that is co-expressed with the UV-induced pilins (UpsA and Ups) as UV exposure does not induce production of this pilus in mutant S. solfataricus strains lacking this ATPase (Fig. 1b) (Fröls et al. 2008).
Yet another S. solfataricus FlaI paralog, BasE, and a co-expressed FlaJ paralog, BasF, are likely involved in the biosynthesis of the putative bindosome discussed above since deletion of the genes encoding BasEF causes an inability of the cells to grow on glucose or arabinose (Zolghadr et al. 2007). However, this deletion does not prevent membrane localization of both binding proteins, signal peptidase processing, glycosylation or binding activity (see below), which demonstrates that there is still much to learn about the role these surface structures play in nutrient uptake. Interestingly, the operon encoding BasEF also contains three small ORFs, basABC, encoding proteins that contain predicted class III signal peptides (Zolghadr et al. 2007). Deletion of these three genes results in slow growth in media containing glucose and arabinose. These FlaFind positives may have an accessory role in bindosome assembly and their function might be comparable to that of the pseudopilins found in bacterial type II secretion systems and type IV pilin assembly machineries (Bleves et al. 1998).
As noted above, gram-negative bacteria have two ATPases, PilB and PilT, that are involved in pilus extension and retraction, respectively. Archaeal homologs have only been identified for PilB (Peabody et al. 2003), indicating that perhaps archaeal pili and flagella are unable to retract. On the other hand, the PilB homologs might provide the necessary energy for both extension and retraction in archaea. Therefore, the structural similarities between the Aquifex aeolicus bacterial PilT and the Archaeoglobus fulgidus archaeal PilB homolog (GspE) are intriguing (Yamagata and Tainer 2007; Forest et al. 2004). Currently, these are the only solved structures available for PilB or PilT.
Despite limited sequence similarities, these PilT and GspE structures have similar domain organization and subunit folding. In the hexameric PilT structure, four domains have open conformations and only a single subunit is clearly bound to an ADP. In the other two subunits, the N-terminal half of the protein twists away from the C-terminal half of the protein. This indicates that these ATPases may be extremely flexible, providing the potential for the leverage to pull pilins out of a pilus. In GspE, all subunits are nucleotide-bound (AMP–PNP), but exist in alternating open–closed conformations. Further studies of GspE have indicated that the N-terminal halves of subunits shift away from the hexameric ring in the open conformation. This provides the potential for significant movement upon ATP hydrolysis, which might promote pilus assembly as well as disassembly. Further insight into the significance of the structural conservation will be obtained from future studies of bacterial PilB structures.
It should be noted that operons encoding flagellins also contain a gene that encodes a second protein that may be an ATPase, FlaH; however, while this potential ATPase contains a Walker A motif, it does not contain a recognizable Walker B motif. Therefore, FlaH hydrolysis of ATP must be demonstrated to confirm that this protein actually is an ATPase. Another possible function for this protein is to control the activity of FlaI. In addition to flagellins, the two ATPases and FlaJ, the flagellin operons encode additional flagella-operon specific accessory proteins; the functions of these proteins, which appear to be archaea-specific, are poorly understood. M. maripaludis deletion mutants that lack lack any of the following: FlaC, FlaF, FlaG, FlaH, FlaI or FlaJ were not motile and did not have flagella, although the flagellin subunits were processed and present in cell membranes (Chaban et al. 2007; Thomas et al. 2002). FlaD and FlaE deletions could not be generated. While archaeal flagella rotate and are likely to drive swimming motility, the mechanisms controlling the rotation of archaeal flagella have not yet been elucidated. Determining and understanding the roles played by these proteins in flagella biosynthesis and/or function, is an exciting prospect (Kupper et al. 1994).
Structures of flagella and type IV pilus-like structures
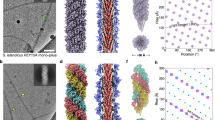
Structural studies of archaeal euryarchaeal and crenarchaeal flagella have shown that like type IV pili, archaeal flagella are thin right-handed helical filaments having central cores comprised of the conserved N-terminal hydrophobic domains of their constituent subunits (Cohen-Krausz and Trachtenberg 2008; Szabó et al. 2007a; Trachtenberg et al. 2005; Trachtenberg and Cohen-Krausz 2006). This is in contrast to bacterial flagella, which have a lumen with a diameter of 2 nm through which flagellins can be transported to the distal tip of the flagellum for assembly (Macnab 2004). While in all solved flagella structures, the exterior domain of the flagellins forms a dominant three-start helix that surrounds the inner core, the outer diameters seem to vary with the Halobacterium salinarium and S. solfataricus flagellum, which is 14 nm in contrast to 11 nm for the Sulfolobus shibatae flagella (Cohen-Krausz and Trachtenberg 2008; Szabó et al. 2007a; Trachtenberg et al. 2005). Interestingly, S. shibatae flagella also have stacked disk packing with C3 symmetry. Both symmetries are present in a single flagellum and result from slight changes in subunit packing (Fig. 2). These small structural changes might enable flagella to act as sensors that allow cells to detect environmental changes. In a recent study in Haloarcula marismortui, it was shown that selected strains contain only one of the two functional flagellins in assembled flagella (Pyatibratov et al. 2008). Interestingly, the flagella formed by FlaA2 are 20–22 nm in diameter, whereas flagella containing only FlaB are 16–18 nm in diameter. This suggests possible flagellin switching, which might be employed to protect cells from hypothetical flagellum-specific haloarchaeal phages.
The three dimensional reconstruction of the S. shibatae flagellum: The filament exhibits two different symmetries. The surface rendered volume (a) and contoured cross-section (d) of the helical symmetry show strong interactions of the outer domains of the subunits. In stacked disk packing with C3 symmetry, there is continuity along the three-start helices in the rendered volume (c), but no contact of the outer domains in the cross-section (f). Both symmetries are found in one filament and connected by the state shown in (b) and (e). With permission from (Cohen-Krausz and Trachtenberg 2008)
Using electron cryo-microscopy, the first archaeal pilus structure was recently solved. Similar to archaeal flagella, this structure, which has been proposed to consist of the EppA substrates (EpdA, B and/or C) (see above) can adopt either of two different symmetries, a C4 symmetry or a one-start helical symmetry that can be present in a single pilus filament; the transitions between symmetries is immediate (Wang et al. 2008). In contrast to archaeal flagella and bacterial type IV pili, this pilus has a central lumen, which is uniform in the portion of the pilus having one-start helical symmetry. The lumen is slightly smaller than that found in bacterial flagella and the type III secretion filament. Further research will be necessary to determine whether the lumen of this pilus is involved in the secretion of pilus subunits or other substances.
The differences between the signal peptides of EppA processed pilin-like proteins and flagellins may partially be responsible for the structural differences between archaeal flagella and this pilus. However, the negatively charged amino acid found at position +5 relative to the peptidase cleavage site in the EppA processed proteins is also found in bacterial type IV pilins, which has a structure that is distinct from both archaeal flagella and this pilus (Craig et al. 2006). Future analysis of archaeal pilus structures will provide more evidence of the diversity of cell surface structures, which may lead to important insights into structure–function relationships.
Functional diversity of type IV pilus-like structures
While type IV pili play important functional roles in several bacterial cellular processes, such as twitching motility, surface adhesion, autoaggregation, conjugation and phage attachment and regress, closely related pilus-like structures, known as pseudopili, are involved in a wide-variety of distinct cellular processes such as protein transport across the outer membrane of gram-negative bacteria (Bradley 1980; Filloux 2004; Hall-Stoodley et al. 2004; Mattick 2002; Merz et al. 2000; O’Toole and Kolter 1998; Vignon et al. 2003).
Moreover, in Geobactre sulfurreducens, which generates energy via electron transfer to extracellular acceptors such as Fe(III) oxides, in addition to the functional roles typically played by type IV pili in processes such as surface adhesion and cell aggregation, type IV pili allow these bacteria to bind and capture a significant quantity of otherwise unavailable electron acceptors by extending the surface area of the cell (Reguera et al. 2005, 2007). Similarly, in natural competence systems of gram-positive bacteria, to efficiently acquire exogenous DNA, small pilin-like proteins and homologs of gram-negative pilus biosynthesis components are involved in assembling cell surface complexes that can bind DNA (Chen et al. 2006; Chung et al. 1998).
Type IV pilus-like structures are also involved in a variety of distinct archaeal cellular processes. For instance, as noted above, archaea are unique in using structures resembling type IV pili to drive swimming motility (Trachtenberg and Cohen-Krausz 2006; Jarrell and McBride 2008). In addition to their primary role in swimming motility, recent studies have revealed that certain archaeal flagella can also facilitate surface attachment. For example, deletions of genes that encode flagellin subunits not only render the crenarcheon S. solfataricus non-motile, they also prevent it from attaching to solid surfaces (Szabó et al. 2007a), unpublished results). Similarly, anti-flagella antibodies inhibit Pyrococcus furiosus binding to several types of abiotic surfaces (Nather et al. 2006). P. furiosus cells can also form connections with each other and establish a bi-species biofilm with Methanopyrus kandleri, processes that may involve P. furiosus flagella (Schopf et al. 2008). However, direct cell–cell interactions between these archaea have also been observed, possibly involving a distinct mechanism. Since FlaFind analyses have identified several genes that encode non-flagellin type IV pilin-like proteins, these close cell–cell interactions may also involve type IV pilus-like structures (Szabó et al. 2007b).
In fact, although UV exposure can induce wild-type S. solfataricus cell–cell interactions, aggregates of these cells are drastically reduced in deletion mutants lacking the above-mentioned UV-induced pilins, UpsA and UpsB, suggesting that type IV pili-like structures are involved in facilitating cell–cell associations as they are in bacteria (Fröls et al. 2008). The genes encoding these proteins are also co-regulated with a helicase, suggesting that autoaggregation may mediate conjugation between UV-exposed cells, possibly facilitating the repair of UV-damaged chromosomal DNA. Interestingly, co-regulation of a helicase with type IV pilin subunits may not be restricted to Sulfolobales strains. Sequence analyses show that operons containing genes encoding FlaFind positives and a helicase are also found in all completed Pyrococcus genome sequences (Szabó et al. 2007b).
Other than flagellins, the largest class of putative proteins identified by the FlaFind program contains putative SBPs (Szabó et al. 2007b). Of the archaea genomes analyzed, the one encoding the largest number of SBPs having predicted class III signal peptides is the crenarchaeon S. solfataricus. It is now known that three of 8 these proteins are involved in the transport of glucose, arabinose and trehalose (Zolghadr et al. 2007). The fact that these subunits have bona fide class III signal peptides and that deletion of the S. solfataricus genes encoding PilB and PilC paralogs impairs the uptake of these sugars strongly suggests that these proteins are involved in the biosynthesis and assembly of an SBP surface structure (Albers et al. 2003; Zolghadr et al. 2007). Similar to bacterial pili and pseudopili that bind substrates (e.g. Fe(III) oxides and DNA—see above), proposed archaeal bindosomes may facilitate the efficient uptake of nutrients by increasing their local concentration. A combination of biochemical and electron microscopic analyses of archaeal cell surfaces should reveal more about archaeal bindosomes that share characteristics with type IV pili.
The majority of genes identified by FlaFind in silico analyses of twenty-two archaeal genomes encode putative proteins not closely related to any previously characterized protein (Szabó et al. 2007b). As previously noted, the organization of an operon can indicate possible functions for the proteins it encodes as is the case for the S. and Pyrococccus operons that contain a gene that encodes a helicase. However, functions for most putative archaeal type IV pilins cannot be easily predicted, due in part to the fact that other genes found in the same operons also encode proteins of unknown function. It is intriguing to note though that archaea often contain many genes encoding very distinctive putative pilin-like subunits and that these putative proteins appear to be differentially regulated, indicating that structures containing these subunits would have different functions. Consistent with this hypothesis, putative Haloferax volcanii type IV pilin-like subunits, which do not share sequence homology beyond the N-terminal signal peptide, are encoded by six differentially regulated operons (Hartman et al. in preparation). These predicted pili may be involved in several distinct biological processes that in bacteria are mediated by type IV pili. Interestingly, surface adhesion, autoaggregation, conjugation and twitching motility all appear to be flagella-independent in Hfx. volcanii (Pohlschröder, unpublished data).
Relatively unique highly conserved domains have been identified in many archaeal pilin-like proteins. For instance, the EppA-recognition site had been identified as a PFAM domain of unknown function (Duf361) in a large number of putative euryarchaeal pilin-like proteins (a putative protein containing this domain has recently been identified in crenarchaea). Hence, as other conserved sequences are also likely to have conserved functions, identifying conserved domains in combination with other in silico, genetic, and biochemical analyses as well as gene expression data and phenotypic data derived from mutant strains may reveal novel structure–function relationships for type IV pilus-like structures.
Conclusions
Despite the fact that only a few detailed studies of archaeal type IV pilus-like filaments have been completed, it is already clear that these structures play important roles in a variety of archaeal cellular processes. Although the core components of bacterial and archaeal type IV pilus biosynthesis machineries appear to be similar, the structures of these archaeal filaments appear to differ substantially from each other and those that have been identified in bacteria. This suggests that further characterization of archaeal type IV pilus-like structures will lead to the identification of novel functional roles for these surface structures. It should also be noted that archaea contain surface filaments that are distinct from type VI pili, such as the recently identified Methanothermobacter thermoautotrophicus pili, composed of subunits that lack class III signal peptides (Thoma et al. 2008). Since in silico and in vivo systems for studying cren- and euryarchaeal genetics are now available, and biochemical methods are well established, a wealth of structural and functional information pertaining to archaeal pilus-like structures should soon be forthcoming. Although, as noted above, the structures and functions of many of these archaeal pili are likely to be unique, studying type IV pili in archaea may also lead to the identification and characterization of novel, related structures in bacteria. Finally, the lack of an outer membrane in archaea and hence a presumably simpler type IV pilus biosynthesis machinery, will facilitate the identification and characterization of components required for the assembly of these structures in all prokaryotes.
References
Albers SV, Szabó Z, Driessen AJM (2003) Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J Bacteriol 185:3918–3925
Albers SV, Szabó Z, Driessen AJM (2006) Protein secretion in the Archaea: multiple paths towards a unique cell surface. Nat Rev Microbiol 4:537–547
Arts J, van Boxtel R, Filloux A, Tommassen J, Koster M (2007) Export of the pseudopilin XcpT of the Pseudomonas aeruginosa type II secretion system via the signal recognition particle-Sec pathway. J Bacteriol 189:2069–2076
Bardy SL, Jarrell KF (2002) FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol Lett 208:53–59
Bardy SL, Jarrell KF (2003) Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol Microbiol 50:1339–1347
Bardy SL, Eichler J, Jarrell KF (2003) Archaeal signal peptides—a comparative survey at the genome level. Protein Sci 12:1833–1843
Bardy SL, Ng SY, Jarrell KF (2004) Recent advances in the structure and assembly of the archaeal flagellum. J Mol Microbiol Biotechnol 7:41–51
Bleves S, Voulhoux R, Michel G, Lazdunski A, Tommassen J, Filloux A (1998) The secretion apparatus of Pseudomonas aeruginosa: identification of a fifth pseudopilin, XcpX (GspK family). Mol Microbiol 27:31–40
Bradley DE (1980) A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can J Microbiol 26:146–154
Burrows LL (2005) Weapons of mass retraction. Mol Microbiol 57:878–888
Chaban B, Ng SY, Jarrell KF (2006a) Archaeal habitats—from the extreme to the ordinary. Can J Microbiol 52:73–116
Chaban B, Voisin S, Kelly J, Logan SM, Jarrell KF (2006b) Identification of genes involved in the biosynthesis and attachment of Methanococcus voltae N-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol Microbiol 61:259–268
Chaban B, Ng SY, Kanbe M, Saltzman I, Nimmo G, Aizawa S, Jarrell KF (2007) Systematic deletion analyses of the fla genes in the flagella operon identify several genes essential for proper assembly and function of flagella in the archaeon, Methanococcus maripaludis. Mol Microbiol 66:596–609
Chen I, Provvedi R, Dubnau D (2006) A macromolecular complex formed by a pilin-like protein in competent Bacillus subtilis. J Biol Chem 281:21720–21727
Chung YS, Breidt F, Dubnau D (1998) Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis. Mol Microbiol 29:905–913
Cohen-Krausz S, Trachtenberg S (2008) The flagellar filament structure of the extreme acidothermophile Sulfolobus shibatae B12 suggests that archaeabacterial flagella have a unique and common symmetry and design. J Mol Biol 375:1113–1124
Conway de Macario E, Macario AJ (2008) Methanogenic archaea in health and disease: a novel paradigm of microbial pathogenesis. Int J Med Microbiol 299:99–108
Correia JD, Jarrell KF (2000) Posttranslational processing of Methanococcus voltae preflagellin by preflagellin peptidases of M. voltae. J Bacteriol 182:855–858
Craig L, Li J (2008) Type IV pili: paradoxes in form and function. Curr Opin Struct Biol 18:267–277
Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA (2006) Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell 23:651–662
Falkowski PG, Fenchel T, Delong EF (2008) The microbial engines that drive Earth’s biogeochemical cycles. Science 320:1034–1039
Filloux A (2004) The underlying mechanisms of type II protein secretion. Biochim Biophys Acta 1694:163–179
Forest KT, Satyshur KA, Worzalla GA, Hansen JK, Herdendorf TJ (2004) The pilus-retraction protein PilT: ultrastructure of the biological assembly. Acta Crystallogr D Biol Crystallogr 60:978–982
Francetic O, Buddelmeijer N, Lewenza S, Kumamoto CA, Pugsley AP (2007) Signal recognition particle-dependent inner membrane targeting of the PulG Pseudopilin component of a type II secretion system. J Bacteriol 189:1783–1793
Francis CA, Beman JM, Kuypers MM (2007) New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation. ISME J 1:19–27
Fröls S, Gordon PM, Panlilio MA, Duggin IG, Bell SD, Sensen CW, Schleper C (2007) Response of the hyperthermophilic archaeon Sulfolobus solfataricus to UV damage. J Bacteriol 189:8708–8718
Fröls S, Ajon M, Wagner M, Teichmann D, Zolghadr B, Folea M, Boekema EJ, Driessen AJ, Schleper C, Albers SV (2008) UV-inducible cellular aggregation of the hyperthermophilic archaeon Sulfolobus solfataricus is mediated by pili formation. Mol Microbiol 70:938–952
Hall-Stoodley L, Costerton JW, Stoodley P (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108
Hansen JK, Forest KT (2006) Type IV pilin structures: insights on shared architecture, fiber assembly, receptor binding and type II secretion. J Mol Microbiol Biotechnol 11:192–207
Jarrell KF, McBride MJ (2008) The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476
Kupper J, Marwan W, Typke D, Grunberg H, Uwer U, Gluch M, Oesterhelt D (1994) The flagellar bundle of Halobacterium salinarium is inserted into a distinct polar cap structure. J Bacteriol 176:5184–5187
Lepp PW, Brinig MM, Ouverney CC, Palm K, Armitage GC, Relman DA (2004) Methanogenic Archaea and human periodontal disease. Proc Natl Acad Sci USA 101:6176–6181
Macnab RM (2004) Type III flagellar protein export and flagellar assembly. Biochim Biophys Acta 1694:207–217
Mattick JS (2002) Type IV pili and twitching motility. Annu Rev Microbiol 56:289–314
Merz AJ, So M, Sheetz MP (2000) Pilus retraction powers bacterial twitching motility. Nature 407:98–102
Nather DJ, Rachel R, Wanner G, Wirth R (2006) Flagella of Pyrococcus furiosus: multifunctional organelles, made for swimming, adhesion to various surfaces, and cell–cell contacts. J Bacteriol 188:6915–6923
Ng SY, Chaban B, VanDyke DJ, Jarrell KF (2007) Archaeal signal peptidases. Microbiology 153:305–314
Ng SY, Zolghadr B, Driessen AJ, Albers SV, Jarrell KF (2008) Cell surface structures of archaea. J Bacteriol 190:6039–6047
O’Toole GA, Kolter R (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30:295–304
Patenge N, Berendes A, Engelhardt H, Schuster SC, Oesterhelt D (2001) The fla gene cluster is involved in the biogenesis of flagella in Halobacterium salinarum. Mol Microbiol 41:653–663
Peabody CR, Chung YJ, Yen MR, Vidal-Ingigliardi D, Pugsley AP, Saier MH Jr (2003) Type II protein secretion and its relationship to bacterial type IV pili and archaeal flagella. Microbiology 149:3051–3072
Pelicic V (2008) Type IV pili: e pluribus unum? Mol Microbiol 68:827–837
Pennisi E (2005) A mouthful of microbes. Science 307:1899–1901
Pohlschroder M, Gimenez MI, Jarrell KF (2005) Protein transport in Archaea: Sec and twin arginine translocation pathways. Curr Opin Microbiol 8:713–719
Proft T, Baker EN (2009) Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci 66:613–635
Pyatibratov MG, Beznosov SN, Rachel R, Tiktopulo EI, Surin AK, Syutkin AS, Fedorov OV (2008) Alternative flagellar filament types in the haloarchaeon Haloarcula marismortui. Can J Microbiol 54:835–844
Reguera G, McCarthy KD, Mehta T, Nicoll JS, Tuominen MT, Lovley DR (2005) Extracellular electron transfer via microbial nanowires. Nature 435:1098–1101
Reguera G, Pollina RB, Nicoll JS, Lovley DR (2007) Possible nonconductive role of Geobacter sulfurreducens pilus nanowires in biofilm formation. J Bacteriol 189:2125–2127
Robertson CE, Harris JK, Spear JR, Pace NR (2005) Phylogenetic diversity and ecology of environmental Archaea. Curr Opin Microbiol 8:638–642
Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I (2007) The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362
Scanlan PD, Shanahan F, Marchesi JR (2008) Human methanogen diversity and incidence in healthy and diseased colonic groups using mcrA gene analysis. BMC Microbiol 8:79
Schopf S, Wanner G, Rachel R, Wirth R (2008) An archaeal bi-species biofilm formed by Pyrococcus furiosus and Methanopyrus kandleri. Arch Microbiol 190:371–377
Szabó Z, Sani M, Groeneveld M, Zolghadr B, Schelert J, Albers SV, Blum P, Boekema EJ, Driessen AJ (2007a) Flagellar motility and structure in the hyperthermoacidophilic archaeon Sulfolobus solfataricus. J Bacteriol 189:4305–4309
Szabó Z, Stahl AO, Albers SV, Kissinger JC, Driessen AJ, Pohlschroder M (2007b) Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases. J Bacteriol 189:772–778
Thoma C, Frank M, Rachel R, Schmid S, Nather D, Wanner G, Wirth R (2008) The Mth60 fimbriae of Methanothermobacter thermoautotrophicus are functional adhesins. Environ Microbiol 10:2785–2795
Thomas NA, Mueller S, Klein A, Jarrell KF (2002) Mutants in flaI and flaJ of the archaeon Methanococcus voltae are deficient in flagellum assembly. Mol Microbiol 46:879–887
Trachtenberg S, Cohen-Krausz S (2006) The archaeabacterial flagellar filament: a bacterial propeller with a pilus-like structure. J Mol Microbiol Biotechnol 11:208–220
Trachtenberg S, Galkin VE, Egelman EH (2005) Refining the structure of the Halobacterium salinarum flagellar filament using the iterative helical real space reconstruction method: insights into polymorphism. J Mol Biol 346:665–676
Vianna ME, Holtgraewe S, Seyfarth I, Conrads G, Horz HP (2008) Quantitative analysis of three hydrogenotrophic microbial groups, methanogenic archaea, sulfate-reducing bacteria, and acetogenic bacteria, within plaque biofilms associated with human periodontal disease. J Bacteriol 190:3779–3785
Vignon G, Kohler R, Larquet E, Giroux S, Prevost MC, Roux P, Pugsley AP (2003) Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J Bacteriol 185:3416–3428
Wang YA, Yu X, Ng SY, Jarrell KF, Egelman EH (2008) The structure of an archaeal pilus. J Mol Biol 381:456–466
Yamagata A, Tainer JA (2007) Hexameric structures of the archaeal secretion ATPase GspE and implications for a universal secretion mechanism. EMBO J 26:878–890
Zolghadr B, Weber S, Szabó Z, Driessen AJ, Albers SV (2007) Identification of a system required for the functional surface localization of sugar binding proteins with class III signal peptides in Sulfolobus solfataricus. Mol Microbiol 64:795–806
Acknowledgments
SVA was supported by a VIDI grant from the Dutch Science Organization (NWO) and intramural funds of the Max Planck Society. MP was supported by a grant from the National Science Foundation (MCB MCB0832292).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Santos.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Albers, SV., Pohlschröder, M. Diversity of archaeal type IV pilin-like structures. Extremophiles 13, 403–410 (2009). https://doi.org/10.1007/s00792-009-0241-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-009-0241-7