Abstract
Several sporozoite proteins have been associated with Plasmodium falciparum cell traversal and hepatocyte invasion, including the cell-traversal protein for ookinetes and sporozoites (CelTOS), and thrombospondin-related sporozoite protein (TRSP). CelTOS and TRSP amino acid sequences have been finely mapped to identify regions specifically binding to HeLa and HepG2 cells, respectively. Three high-activity binding peptides (HABPs) were found in CelTOS and one HABP was found in TRSP, all of them having high α-helical structure content. These HABPs’ specific binding was sensitive to HeLa and HepG2 cells’ pre-treatment with heparinase I and chondroitinase ABC. Despite their similarity at three-dimensional (3D) structural level, TRSP and TRAP HABPs located in the TSR domain did not compete for the same binding sites. CelTOS and TRSP HABPs were used as a template for designing modified sequences to then be assessed in the Aotus monkey experimental model. Antibodies directed against these modified HABPs were able to recognize both the native parasite protein by immunofluorescence assay and the recombinant protein (expressed in Escherichia coli) by Western blot and ELISA assays. The results suggested that these modified HABPs could be promising targets in designing a fully effective, antimalarial vaccine.
Similar content being viewed by others
Introduction
Highly specialized invasive forms of Plasmodium falciparum (the parasite causing the most lethal form of malaria) recognize, invade, and infect two target cells in human hosts: hepatocytes and red blood cells (RBC) (Kappe et al. 2004; Garcia et al. 2006; Cowman and Crabb 2006). P. falciparum sporozoites (larvae-like structures) injected into the skin during the bite of an infected female Anopheles mosquito travel through the bloodstream to the liver during the first phase of human malaria infection where they cross the sinusoidal layer through Disse’s space and Kupffer cells to invade their primary target: the hepatic cell (Sinnis and Coppi 2007). Two highly relevant proteins participating in such host–pathogen interactions have been widely studied: the circumsporozoite protein (CSP) and the thrombospondin-related anonymous protein (TRAP) (Akhouri et al. 2008; Rathore et al. 2003); both have been shown to mediate sporozoite motility, host-cell recognition, cell traversal, binding, and entry to host cells. These proteins thus constitute targets for intensive research in designing vaccines acting against this deadly disease’s pre-erythrocyte stage (Bermudez et al. 2008; Bongfen et al. 2009; Cifuentes et al. 2008; Khusmith et al. 1991; Kumar et al. 2006). Several studies have shown that sporozoite proteins promote the malaria parasite crossing the dermis and the liver’s sinusoidal wall prior to invading the hepatocytes; SPECT-1 and SPECT-2 are two such sporozoite microneme proteins as they are essential for cell traversal and are expressed in the micronemes and then translocated to the sporozoite surface (Ishino et al. 2004, 2005; Kaiser et al. 2004a; Yuda and Ishino 2004).
Knowledge regarding sporozoite biology, chemistry, and function during this first invasion phase has been limited for several years, mainly due to the small number of sporozoite and pre-erythrocyte forms available, as well as a lack of an in vitro system for producing sporozoites. A large number of attractive molecules from each parasite stage have recently been identified due to the complete P. falciparum genome, proteome, and transcriptome having been analyzed (Florens et al. 2002; Gardner et al. 2002; Kaiser et al. 2004b). These molecules are currently being studied as alternatives for developing more efficient malaria control mechanisms.
Two novel sporozoite proteins from among this promising array have been recently associated with sporozoite cell traversal via liver macrophages (Kupffer) and hepatic cell recognition and infection, namely the cell-traversal protein for ookinetes and sporozoites (CelTOS) and the thrombospondin-related sporozoite protein (TRSP) (Kaiser et al. 2004b; Kariu et al. 2006; Labaied et al. 2007).
CelTOS is translocated to the sporozoite surface and then mediates parasite infectiveness. It is a 25-kDa protein which is expressed in micronemes from both mosquito midgut and mammalian liver-infective sporozoites (Kariu et al. 2006). Targeted disruption of the celtos gene has demonstrated in vitro this protein product’s direct participation in parasite transversal to HeLa cells and cellular barriers in both mosquitoes and vertebrates (Kariu et al. 2006), since celtos-null sporozoites have been shown to have reduced infectivity in mice, whilst in vitro experiments have shown that cell-passage ability becomes almost abolished. Interestingly, sporozoite infectivity was restored in Kupffer cell-depleted rats, suggesting that CelTOS main function is specifically related to sporozoite host-cell traversal ability, possibly by interacting with host-cell molecules on the membrane (Kariu et al. 2006). Interestingly, it has been reported that immunization with Pf CelTOS elicits protection in mice against heterologous challenge with Plasmodium berghei and that CelTOS-specific antibodies can inhibit P. falciparum sporozoite invasion of hepatocytes in vitro and also P. falciparum sporozoite motility in vitro (Bergmann-Leitner et al. 2010), thereby highlighting CelTOS’ potential as a promising vaccine candidate.
The TRSP protein was identified in differential transcriptome analysis of Plasmodium yoelii sporozoites (Kaiser et al. 2004b) as being a 18-kDa protein (163 amino acids long), containing a characteristic signal sequence within its primary structure, a C-terminal hydrophobic region which could serve as a membrane anchor domain (Labaied et al. 2007) and a thrombospondin type 1 repeat (TSR) domain. This TSR domain is characteristic of the P. falciparum TRAP protein family which has been found in several surface proteins involved in ookinete and sporozoite motility, as well as in host cell binding and invasion (Kaiser et al. 2004b; Pradel et al. 2002; Tucker 2004). TRSP has a unique distribution pattern by contrast with other micronemes and surface proteins, suggesting that TRSP is located in the sporozoite rhoptries. Unfortunately, no sporozoite rhoptry-specific proteins have been identified so far, thus precluding co-localization studies confirming this hypothesis (Kaiser et al. 2004b; Labaied et al. 2007). Interestingly, in vitro and in vivo knockout assays with the Plasmodium berghei TRSP homolog have indicated an important role for this protein in hepatocyte invasion; this would also reinforce this protein’s potential as an antimalarial vaccine component (Labaied et al. 2007).
Identifying specific Plasmodium protein-derived antigens participating in host–pathogen interactions in all parasite stages is a highly relevant step for ensuring a logical and rational development strategy for designing a fully effective antimalarial vaccine (Patarroyo et al. 2008a; Cowman et al. 2002). A vaccine is urgently needed for protecting around 3.2 billion people living in high-risk areas, as well as preventing the 300 million cases, and more than 2 million deaths caused by this disease every year (Snow et al. 2005; Hay et al. 2010).
Entire sequences have been synthesized as short synthetic peptides (20-mer-long) regarding CelTOS’ relevance for the necessary cell passage for crossing the sinusoidal layer and TRSP during hepatocyte invasion; each peptide’s ability to bind to HeLa (in vitro model for parasite cell-traversal ability) or HepG2 cells (hepatic cell line for sporozoite invasion) has also been assessed. High-activity binding peptides (HABPs) have thus been identified by using a robust, highly specific and sensitive methodology which has been tailor-made for this purpose. Specific binding has been determined in binding assays by using radiolabeled and non-radiolabeled peptide. The data have been analyzed by using bimolecular interaction theory which has led to finding that high-affinity binding regions recognize around 2,000 binding sites per cell and that they have nanomolar dissociation constants (Patarroyo and Patarroyo 2008; Rodriguez et al. 2008).
This has led to identifying and characterizing a new set of minimal subunit-based, chemically synthesized sporozoite peptides mediating CelTOS interaction with HeLa cells, as well as TRSP interaction with HepG2 cells, in turn, constituting interesting targets for blocking sporozoite invasion during cell-traversal and hepatic infection stages as components of a chemically synthesized, multi-epitope, multistage, minimal subunit-based, fully effective anti-malarial vaccine.
Materials and methods
Peptide synthesis and radiolabeling
The t-Boc strategy was used for synthesizing 20-mer-long (Merrifield 1963; Houghten 1985), non-overlapping peptides from P. falciparum 3D7 strain CelTOS (PFL0800c), and TRSP (PFA0200w) protein sequences, taken from the PlasmoDB database (http://plasmodb.org/plasmo/). Synthesized peptides were named according to our institute’s sequential numbering system (Fig. 1). A tyrosine residue was added to the C-terminal of those peptides which did not contain this residue in their sequence to enable radiolabeling.
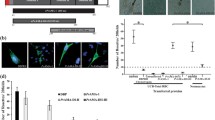
Receptor-ligand assays. Each peptide’s binding activity is represented by the length of the black bars shown in front of each amino acid sequence: a CelTOS and b TRSP. Peptides presenting a ≥2% binding activity cut-off point were considered to be HABPs. CelTOS and TRSP schematic representation, indicating signal sequence position (SS), transmembrane domains (Tm), and thrombospondin-related type I domain (TSR). Two cysteine residues were replaced by threonine (black rectangles) in TRSP peptides 36823 and 36824 due to polymerization issues during peptide synthesis. Conserved protein regions are shown in green and variable regions are shown in yellow. c Total, non-specific, and specific binding for peptide 34452. The slope from trend line in specific binding indicates 3.1% specific binding, meaning that peptide 34452 is a HABP. cpm: counts per minute. d HABP 34451 saturation curve. Increasing amounts of radio-labeled peptide were added in the presence or absence of unlabeled peptide. The curve represents specific binding. In the Hill plot (interior), the abscissa is log F and the ordinate is log(B/B max − B), F being free peptide, B the amount of bond peptide and B max the maximum amount of bound peptide. B max is used for calculating K d and number of sites per cell (Attie and Raines 1995; Rodriguez et al. 2008). e The effect of HeLa and HepG2 cell treatment with heparinase I (HI), heparinase II (HII), chondroitinase (CABC), and chondroitinase AC (CAC) on CelTOS and TRSP HABP binding activity. Peptides binding to untreated cells were used as binding (100%) control (C)
Regarding immunization studies, peptide polymers were obtained after CG had been added to the N- and C-termini to allow peptide polymerization. HPLC-purified peptides were incubated for 15 min with 5 μL Na125I (100 mCi/mL, MP Biomedicals) and 15 μL chloramine-T (2.75 mg/mL) for radiolabeling (Curtidor et al. 2008b). The reaction was stopped by adding 15 μL sodium metabisulfite (2.25 mg/mL). Radiolabeled peptides were purified on a Sephadex G-10 column and analyzed in an Auto Gamma Counter Cobra II (Packard).
HepG2 and HeLa cells
HeLa cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (Gibco) and antibiotic/antimycotic mixture (Gibco). Likewise, HepG2 cells were cultured in DMEM medium supplemented with non-essential amino acids (Gibco) and bovine pancreas insulin (Sigma). Cells were incubated at 37°C in a 5% CO2 atmosphere. After a confluent layer became formed, cells were dissociated using 0.05% EDTA–PBS. Before being used, cells were collected by adding EDTA–PBS and centrifuging; they were then washed with incomplete medium and their viability and concentration were assessed in a Neubauer chamber using trypan blue staining.
CelTOS and TRSP peptide binding assays
HeLa and HepG2 cell binding assays were conducted in triplicate, as described elsewhere (Curtidor et al. 2008b; Lopez et al. 2001). Briefly, increasing concentrations (0–560 nM) of radiolabeled CelTOS- and TRSP-derived synthetic peptides were incubated for 1 h with HeLa or HepG2 cells (1.2 × 106 cells), respectively, in the absence (total binding) or presence (unspecific binding, 140-fold excess) of unlabeled peptide. Cells were recovered from the reaction mixture by spinning at 8,000×g for 5 min through a 60:40 dioctyl phthalate/dibutyl phthalate cushion (1.015 g/mL) and cell-associated radioactivity was quantified by gamma counter. HABPs were defined as being peptides having a ≥0.02 ratio (2% specific binding) according to previously established criteria, keeping in mind that specific binding activity (specific binding activity = total binding − unspecific binding) at four increasing logarithmic concentrations defines the specifically bound peptide (pmol) per added peptide (pmol) ratio (Curtidor et al. 2008b; Rodriguez et al. 2008).
Modified binding assays involving a wider range of concentrations (0–3,200 nM) were carried out to determine the HABP binding equilibrium constants for the interactions established between CelTOS and TRSP HABPs with HepG2 and HeLa cells, respectively. Samples were incubated and separated from the mixture reaction (the same used in binding assays); cell-associated radioactivity was analyzed by gamma counter. The experimental data so obtained was analyzed using saturation function (bound ligand concentration compared with total and or free ligand); Hill coefficients (cooperativity), dissociation constants (K d), and number of sites per cell could thus be obtained (Attie and Raines 1995; Rodriguez et al. 2008).
Binding to enzyme-treated cells and cross-competition assays
Each cell line was suspended in HBS buffer and treated independently for 1 h with 500 μU/mL of heparinase I (HI; CAS 9025-39-2, Sigma), heparinase II (HII; CAS 149371-12-0, Sigma), chondroitinase AC (CAC; CAS 9047-57-8, Sigma), and chondroitinase ABC (CABC; CAS 9024-13-9, Sigma) at 37°C. Treated cells were then washed and assessed in the same way as conventional binding assays, using untreated cells as positive control.
Cross-competition assays were also carried out between TRSP HABP and a TRAP-derived peptide. Briefly, 24 nM of 125I-labeled-HABP 36075 was incubated for 90 min with HepG2 cells (1 × 106) in the presence of unlabeled TRAP HABP 3289 at three concentrations (6, 20 and 80 μM). Cells were then washed before cell-bound radiolabeled peptides were quantified (as described above).
Circular dichroism (CD) and nuclear magnetic resonance (NMR) spectroscopy, and structural calculation
CelTOS and TRSP HABP secondary structures were analyzed by CD. HPLC-purified peptides’ spectra were recorded at 20°C in 30% v/v trifluoroethanol (TFE), using a 1-cm optical path length thermostated quartz cell. All spectra were acquired in a Jasco J-810 equipment (JASCO Inc.) by averaging three sweeps taken at 20 nm/min. Data were processed by Spectra Manager software and analyzed with CONTINLL, SELCON and CDSSTR deconvolution software (Sreerama and Woody 2000).
For NMR experiments, ten milligrams of HPLC purified HABP 36075 was dissolved in 500 μL 30% v/v TFE (Roccatano et al. 2002). Resonance assignments were obtained from two-dimensional TOCSY, DQF-COSY, and NOESY spectra and sequences were assigned following standard procedure (Wüthrich New York 1986). All experiments were carried out using a Bruker DRX-500 MHz spectrometer at 295 K.
Accelrys software was used for determining HABP 36075 structure. NOE peaks (selected from NOESY data sets obtained at 400 ms) were integrated and converted into distance restraints; such restraints were grouped as being strong, medium, and weak, corresponding to 1.8–2.5, 2.5–3.5, and 3.5–5.0 Å distance restraints, respectively. Hydrogen bond constraints were introduced for slow peptide NH exchange rate; distance ranges involving these likely NH–O hydrogen bonds were set at 1.8–2.5 Å. Havel and Wuthrich’s DGII distance geometry software was used for producing 50 starting structures (Havel and Wuthrich 1985).
Aotus monkey immunization with CelTOS and TRSP modified HABPs
CelTOS-derived HABPs 34451 and 34458 and TRSP-derived HABP 36075 were used as templates for designing analog peptides 38138 (CG21NVHTFRGDNVHNSSSSL40GC), 38140 (CG161 IWNYNSDDVSESEESLSDDF180GC) and 38148 (CG41SDVRYNKSFINNRLLNEHAH60GC), respectively, following thoroughly described physicochemical principles (Patarroyo and Patarroyo 2008). Please note that modified peptides and modified residues are written in italics and highlighted in bold.
Analog peptides were used for subcutaneously immunizing groups of 5–8 Aotus monkeys which were kept in our field station in Leticia (Amazonas, Colombia); animal care and handling were in line with Colombian Institute of Health guidelines which was strictly supervised by the competent Colombian environmental authority (CORPOAMAZONIA), as previously described (Curtidor et al. 2007; Cifuentes et al. 2009). The immunization scheme was as follows: 125 μg peptide dissolved in distilled water was homogenized with 250 μL Freund’s complete adjuvant (FCA) for the first dose administered on day zero and with 250 μL Freund’s incomplete adjuvant (FIA) for the second and third doses administered on days 20 and 40, respectively. Each monkey was immunized with 200 μL of the peptide emulsion and control monkeys were immunized with saline solution and CFA or FIA on the same days. Blood was obtained from the femoral vein.
Extracting P. falciparum genomic DNA and PCR amplification
Human RBCs (200 μL) parasitized with either P. falciparum FCB-2 (Colombia), or PAS-2 (unknown origin) or FVO (Vietnam) strains (30% parasitemia) were obtained from an asynchronous culture, maintained as described elsewhere (Lambros and Vanderberg 1979). Erythrocytes were lysed afterwards using 0.2% saponin and genomic DNA (gDNA) from each strain was extracted using an UltraClean DNA blood isolation kit (MO BIO, Carlsbad, CA).
Genes encoding CelTOS and TRSP proteins in the P. falciparum 3D7 reference strain (PFL0800c and PFA0200w, respectively) were analyzed for designing specific primer sets for amplifying HABP-encoding regions. A single primer set was designed for each protein using Gene Runner v3.05; sequences were CelTOS-F (5′-CGTATTACGTTTGTTTGTTTG-3′) and CelTOS-R (5′-AAATTAGCACACACATATATAC-3′) amplifying the region encoding HABPs 34451, 34452 and 34458, as well as TRSP-F (5′-GGCTTTATCCGTTCACGAC-3′), and TRSP-R (5′-TGATCTGTGCTTATTTTTACTC-3′) amplifying the HABP 36075-encoding region. The HABP 33577-encoding region in P. falciparum integral membrane protein Pf25-IMP was included as a positive PCR control; it was amplified using DIR1 and REV1 primers (Curtidor et al. 2008a).
DNA regions were amplified in 50 μL reaction mixture using 1.25U BioTaqTM DNA polymerase (Bioline, London, UK). The following thermocycling profile was used for all primer sets: an initial denaturing step at 95°C for 5 min, followed by 35 cycles consisting of: 1 min of annealing at 56°C, 1 min of extension at 72°C, and 1 min denaturing at 95°C, followed by a final extension cycle at 72°C for 5 min. The same reaction conditions were established using DNase- and RNase-free water instead of DNA as negative control. Amplification products were purified using a Wizard PCR preps kit (Promega, Madison, WI) and sequenced using their corresponding forward and reverse primers.
Recombinant protein cloning and expression
The P. falciparum FCB-2 strain CelTOS (residues 25–182) and TRSP (residues 39–138) putative protein encoding regions were amplified from genomic DNA using the following specific primers: forward-CelTOS: 5′-ATGTTCAGAGGAAACAACGGA-′3, reverse-CelTOS: 5′-ATCGAAAAAATCATCTGATA-3′, forward-TRSP: 5′-ATGCTTATGAAAATTTCAAG-3′ and reverse-TRSP: 5′-GATTAAAATATATGTTGTTCG-3′. PCR products were cloned in pEXP5-CT/TOPO expression vector (Invitrogen) which adds a polyhistidine (6-His) tag at proteins’ C-terminus to facilitate further purification and detection. Recombinant plasmid integrity was corroborated by an ABI PRISM 310 automatic genetic analyzer (PE Applied Biosystems). ClustalW was used for comparing FCB2 strain sequencing results with those for the 3D7 reference strain through nucleotide and amino acid alignment (Thompson et al. 2002).
Recombinant proteins were expressed in E. coli BL21-AI cells after being induced with 0.2% l-arabinose. Harvested cell pellets were solubilized in denaturing lysis buffer (6 M Urea, 10 mM sodium phosphate, 10 mM Tris–Cl, 15 mM Imidazole) supplemented with protease inhibitors (100 mM PMSF, 0.5 M EDTA, 1 mg/mL leupeptin, 100 mM iodoacetamide) and 1 mg/mL lysozyme. The clear supernatant was applied to a pre-equilibrated Ni–NTA agarose column (Qiagen) to purify the recombinant protein by solid-phase affinity chromatography, as has been previously described (Mongui et al. 2010). The presence of both CelTOS (rPfCelTOS) and TRSP recombinant proteins (rPfTRSP) was verified by SDS–PAGE and Western blot (WB) using peroxidase-coupled monoclonal anti-polyhistidine antibodies (Mongui et al. 2010). Recombinant proteins were thoroughly dialyzed against PBS pH 7.5 and the amount of protein was determined by Micro BCA Protein Assay kit (Thermo Scientific).
Enzyme-linked immunosorbent assay (ELISA) with recombinant proteins
rPfCelTOS and rPfTRSP recognition and antibody titers were determined by ELISA. In brief, 96-well plates coated with 5 μg/mL rPfCelTOS or rPfTRSP were incubated with modified peptide-immunized Aotus monkey sera (1:100 initial dilution), followed by twofold serial dilutions in the entire row of wells. Anti-CelTOS sera and anti-TRSP sera final dilutions were 1:6,400 and 1:3,200, respectively. Peroxidase-coupled anti-Aotus IgG (1:10,000) was used as secondary antibody and immunoreactivity was revealed using a TMB Microwell peroxidase substrate system kit (KPL Laboratories, WA, USA), according to the manufacturer’s instructions. Aotus monkey antibody titers were determined by successive twofold dilutions of primary antibody, until reaching an A450 value equal to pre-immune sera value ± 2SD.
Indirect immunofluorescence assays (IFA) and WB analysis
P. falciparum 3D7 strain sporozoite air-dried slides, fixed with 2% BSA in PBS (kindly provided by Dr. Patricia de la Vega) were used for assessing the reactivity of Aotus monkey sera immunized with CelTOS (38138, 38140) and TRSP (38148) analog peptides. Slides were incubated for 30 min with monkey sera at 1:40 dilution. Pre-immune sera from all monkeys were used as negative controls. Fluorescence was observed using the F(ab)2 fragment from affinity purified goat anti-monkey IgG:rhodamine isothiocyanate (TRITC) conjugate in 1:100 dilution (appearing as red by fluorescence microscopy). Anti-CSP serum produced in an Aotus monkey was used as primary antibody at a 1:100 dilution for CelTOS co-localization studies and then detected with goat anti-Aotus IgG conjugated to fluorescein isothiocyanate (FITC) at a 1:100 dilution (appearing as green).
rPfCelTOS or rPfTRSP was separated by 12% SDS–PAGE in non-reducing conditions and then transferred to a nitrocellulose membrane for WB analysis. Membranes were blocked with 5% skimmed milk in PBS-0.05% Tween and washed thrice with PBS-0.05% Tween. Nitrocellulose strips were individually incubated with monkey sera diluted 1:100 in blocking solution, washed several times, incubated with goat anti-Aotus IgG conjugated to alkaline phosphatase (AP) at 1:1000 dilution, and finally developed with NBT/BCIP (Vector Laboratories).
Results
CelTOS and TRSP peptides interacted with HeLa and HepG2 cells, respectively
Three HABPs were found in CelTOS which had high specific HeLa cell binding activity and affinity: 34451 (21NVLCFRGNNGHNSSSSLYNG40), 34452 (41SQFIEQLNNSFTSAFLESQSY 60) and 34458 (161IWNYNSPDVSESEESLSDDF180). The first two were located in the CelTOS N-terminal region and the latter was located towards its C-terminal (Fig. 1a). Only one HepG2 cell-binding HABP was identified in TRSP, namely 36075 (41SDVRYNKSFINNRLLNEHAH60), which was located towards its TSR domain’s N-terminal region (Fig. 1b). Figure 1c shows as an example of the plots obtained for peptide 34452 total, non-specific and specific binding. The specific binding plot slope (0.311) indicated 3.1% specific binding, meaning that peptide 34452 was considered to be a HABP (Fig. 1a).
CelTOS HABPs had high-affinity HeLa cell interactions, as shown by their dissociation constants (K d) which came within the nanomolar range (500 nM for HABP 34451 and 680 nM for HABP 34458) and recognized around 7,200,000 (HABP 34451) and 139,000 (HABP 34458) sites per cell. Hill coefficients (n H) obtained from saturation curves were higher than 1 for both HABPs (1.18 for HABP 34451 and 1.90 for HABP 34458), suggesting these peptides’ positive cooperative effect on CelTOS-mediated cell-traversal activity. Figure 1d shows HABP 34451 saturation curve and Hill plot. CelTOS HABP 34452 and TRSP HABP 36075 binding did not reach saturation in the conditions being assessed here, due perhaps to a larger number of binding sites per cell for these peptides.
CelTOS and TRSP HABPs bound differentially to enzyme-treated HeLa and HepG2 cells
HI-, HII-, CAC- and CABC-treated HepG2, and HeLa cells weakly affected CelTOS and TRSP HABP binding. TRSP HABP 36075 HepG2 cell binding was weakly affected by HI treatment, whereas CelTOS HABP 34451 and 34452 HeLa cell binding was slightly sensitive to CABC (less than 24% for each HABP) (Fig. 1e).
CelTOS and TRSP HABP polymorphism assessed in different strains
CelTOS and TRSP HABP-encoding regions from FCB-2, PAS-2 and FVO strains were amplified by PCR, as described above. Two different sized amplification products (850 and 291 bp) were observed for CelTOS and TRSP, respectively, agreeing with expected sizes. The control primers amplified a single band of around 438 bp (Curtidor et al. 2008a) (Fig. 2a).
Molecular biology assays. a Lanes 1, 2, and 3, PCR amplification of the region encoding FCB-2, FVO and PAS-2 strain gDNA CelTOS HABPs 34451, 34452, and 34458, respectively. Lanes 4, 5, and 6: amplification of FCB-2, FVO, and PAS-2 strain gDNA TRSP HABP 36075, respectively. Lanes 7, 8, and 9: positive control (Pf25-IMP) for the FCB-2, FVO and PAS-2 strains, respectively. NC: PCR negative control. b CelTOS (top) and TRSP (down) amino acid sequence alignment from different P. falciparum strains using MEGA 4 software (Tamura et al. 2007). FCB-2, FVO and PAS-2 strains were sequenced and compared with other strains reported in Broad Institute projects (http://www.broadinstitute.org/), as well as in the PlasmoDB database (http://plasmodb.org/plasmo/). HABPs are enclosed within black boxes. c PCR amplification of genes encoding P. falciparum CelTOS and TRSP in the FCB-2 strain. MWM: molecular weight marker. Lane (1) CelTOS (left) and TRSP (right), PCR of genomic DNA. Lane (2) negative control. d SDS–PAGE and Western blot of P. falciparum CelTOS (left) and TRSP (right) purified proteins. Lane (1) non-induced bacterial lysate. Lane (2) l-arabinose-induced bacterial lysate. Lane (3) SDS–PAGE of purified protein. Lane (4) Western blot of non-induced lysate and Lane (5) Western blot of purified protein using anti-polyhistidine monoclonal antibody
CelTOS nucleotide and amino acid sequences from the three aforementioned P. falciparum strains were aligned with those from the 3D7 (Airport malaria, Amsterdam, The Netherlands) and Dd2 (Thailand) reference strains, while TRSP sequences were only compared with the 3D7 reference strain; ClustalW software was used for all alignments (Combet et al. 2000). The CelTOS sequence had 12 nucleotide changes (8 single and 2 double non-synonymous substitutions, therefore producing a shift in 10 amino acid residues). Strain-specific polymorphisms in HABP regions described below, with their positions, have been numbered regarding the 3D7 reference strain. T to G nucleotide substitution was observed in HABP 34451 N-terminus in position 63 in the Dd2 strain, shifting asparagine to lysine in residue 21. Four nucleotide substitutions were found in HABP 34458-encoding sequences: one T to A substitution in position 496, shifting serine (S) to threonine (T) in residue 166 in FCB-2, PAS-2 and FVO strains, (2) a double substitution in nucleotide positions 499 (from T to C) and 500 (from C to T), shifting proline (P) to leucine (L) in residue 167 in the Dd2 strain and to serine (S) in the FCB-2, PAS-2 and FVO strains, (3) a T to C mutation in position 508, shifting serine (S) to proline (P) in residue 170 in FCB-2, PAS-2 and FVO and (4) a G to A substitution was found in position 524, shifting (S) to asparagine (N) in residue 175 in the Dd2, FCB-2, PAS-2, and FVO strains. The other substitutions were found outside HABP-encoding sequences (Fig. 2b).
Amino acid sequences obtained from the three P. falciparum strains for TRSP HABP 36075 were seen to be 100% identical to the reference sequence; no substitutions were found in the nucleotide sequence (Fig. 2b).
Recombinant protein production
celtos and trsp gene amplification revealed specific ~474 bp and ~300 bp bands, respectively (Fig. 2c). Recombinant protein expression was observed after l-arabinose induction, a single ~17 kDa band being detected with rPfCelTOS anti-polyhistidine monoclonal antibody (Fig. 2d), thereby agreeing with this protein’s predicted molecular weight without signal peptide (Kariu et al. 2006). The anti-polyhistidine monoclonal antibody detected a single specific ~12 kDa band (Fig. 2d) for rPfTRSP, in agreement with the expected molecular weight for the recombinant protein fragment so obtained. Proteins were purified and dialyzed against PBS to ensure that denaturing agents had been completely removed and also allow protein folding.
TRAP HABP 3289 did not compete for HABP 36075 binding sites
Cross-competition assays with TRSP HABP 36075 and previously-identified TRAP HABP 3289 (Lopez et al. 2001) showed that TRAP HABP did not specifically compete for TRSP HABP 36075 binding sites when using different competitor peptide concentrations.
CelTOS and TRSP HABPs had similar secondary structure features
High α-helix content in CelTOS 34451, 34452, and TRSP 36075 HABP structures was determined by CD spectroscopy; two characteristic minima were observed in all spectra at 206 and 221 nm and one maximum at 190 nm (Fig. 3a). Spectra deconvolution with SELCON, CONTINLL and CDSSTR software showed greater than 80% α-helix content in agreement with these results. By contrast, HABP 34458 mostly presented 200 nm minimum random elements, agreeing with the low α-helical element content obtained in CD deconvolution results.
CelTOS and TRSP HABP structural characteristics. a Circular dichroism spectra for CelTOS HABPs 34451, 34452 and 34458, and TRSP HABP 36075, recorded in 30% TFE. b Summary of sequential and medium-range NOE connectivity for HABP 36075, represented by line thickness. Temperature coefficient values less than 4.0 used in the calculation are indicate as asterik. c Ribbon representation of HABP 36075 determined by NMR is shown in red. The 3D structure for the TRS domain in TRAP protein (PDB 2BBX) is shown in silver, while regions where peptides 3287 and 3289 sequences are located are shown in blue and orange, respectively. At the bottom, backbone representation from HABP 3289 (orange) and 36075 (red) 3D structures (determined by NMR) superimposed on PfTRAP-TSR β-turn 258GKGT261 fragment backbone (blue) (PDB 2BBX)
NMR HABP 36075 structure determination
NOESY spectra showed that dαN (i, i + 1) sequence signals were stronger than intra-residue cross-peaks. The presence of dNN cross-peaks indicated a significant population of conformations in the α-region of ΦΨ space, as well as some medium-range dαβ (i, i + 3), dαN (i, i + 3), and dαN (i, i + 4) NOE connectivity, suggesting the presence of an α-helical structure between residues N11–H20 and a helical trend between K7-I10 (Fig. 3b).
Twenty-four modeled structures (from the original 50) whose distance violations were no greater than 0.25 Å and whose ω angles were greater than 1.5° were chosen. An average 0.24 Å RMSD (Root Mean Square Deviation) was obtained for the main-chain atoms by superimposing structures between residues N6-H20 with a consensus structure having the lowest total energy. Secondary structure analysis showed the presence of an α-helix between residues N11-H20; this structural feature was confirmed by medium-range NOEs and by dihedral Φ and Ψ angles for each residue in the helical region, adopting equal values (approximately −60° and 45°, respectively). A helical trend was observed between residues K7-I10 (Fig. 3c).
Immunoassays
Recognizing native protein by anti-HABP modified antibodies
One monkey out of eight immunized with analog peptide 38138 (34451) and three more monkeys immunized with analog 38140 (34458) developed specific antibodies which strongly reacted with small sporozoite intracytoplasmic structures by immunofluorescence (Fig. 4ai, aii, red), suggesting microneme location, as has been previously reported for this protein. The reactivity obtained with the Aotus anti-CSP molecule displayed a green fluorescence pattern on the periphery, suggesting a membrane location (Fig. 4ai, aii, green).
Immunological assays. a Aotus monkey sera immunized with modified HABPs recognized CelTOS and TRSP in P. falciparum sporozoites by immunofluorescence. A dual indirect fluorescence assay carried out with antisera produced in Aotus monkeys (specific for CSP, gives green fluorescence) showed microneme labeling (small dots) and also the protein’s presence on membrane. Antibodies against CelTOS-derived analog peptides (i) 38138 and (ii) 38140 revealed small intracytoplasmic dots similar to micronemes. (iii) TRSP protein detection with antibodies against analog peptide 38148, showing an internal bilobed staining pattern. b Immunized monkeys’ antibody titers were determined using ELISA with CelTOS analog peptides 38138 and 38140, and TRSP 38148. Two monkeys’ sera dilutions starting at 1:100 are shown, until reaching the target’s absorbance ±2SD. Each line represents a monkey assessed here. c Western blot assays. Left panel: rPfCelTOS recognition Lane 1–4: Pre-immune serum. Lane 5: Hyper-immune monkey serum immunized with 38138 peptide analog. Lane 6–8: hyper-immune monkey sera immunized with analog peptide 38140. Right panel: Lanes 1–6: Pre-immune serum. Lanes 7–12: recognition of rPfTRSP protein by monkey sera immunized with 38148 peptide analog
Aotus antibodies raised against TRSP analog peptide 38148 (36075) revealed an internal bilobulated staining pattern, suggesting its location in sporozoite rhoptries; however, sporozoite rhoptry-specific proteins have not been reported to date (Fig. 4aiii). Unfortunately, double-staining with anti-CSP and TRSP was not carried out because of the scarcity of sporozoite slides.
Antibodies against CelTOS and TRSP protein analog peptides recognized recombinant proteins by ELISA and WB
Specific antibody titers against rPfCelTOS and rPfTRSP were obtained by successive dilutions of monkey sera immunized with analog peptides 38138 (34451), 38140 (34458), and 38148 (36075); 1:400 titers were found for the single monkey immunized with peptide 38138 and titers ranging from 1:800 to 1:6,400 were found for monkeys immunized with peptide 38140 (Fig. 4b). Titers ranging from 1:800 to 1:3,200 for TRSP modified HABP 38148 (36075) were found for all six monkeys assessed in this study (Fig. 4b, right).
A ~17 kDa band was detected by WB when the same monkey sera inoculated with CelTOS analog peptides 38138 (34451) and 38140 (34458) was incubated with rPfCelTOS (Fig. 4c). ~36 kDa and ~54 kDa bands were detected in some cases which could have corresponded to the dimerized and trimerized recombinant protein; this resulted from a lack of reduction/alkylation treatment of the sample prior to electrophoresis. Likewise, monkey sera inoculated with TRSP analog peptide 38148 (36075) recognized a specific ~12 kDa band and a very weak ~27 kDa band which could have corresponded to the dimerized recombinant protein (Fig. 4c).
Discussion
Hepatocyte invasion by Plasmodium falciparum sporozoites represents the initial stage in the most lethal form of human malaria. Sporozoites must overcome natural barriers to reach their host/target cell (the hepatocyte), including circumventing the liver’s sinusoidal barrier and Disse’s space via Kupffer cells (Sinnis and Coppi 2007). Along with CSP, TRAP and SPECT 1 and 2, two new proteins have been recently described as they are directly involved in sporozoite cell-traversal and hepatic cell invasion: CelTOS and TRSP, respectively (Kaiser et al. 2004b; Kariu et al. 2006; Labaied et al. 2007; Bergmann-Leitner et al. 2010). These new proteins represent attractive targets to be used as templates for designing a multi-stage, multiepitope, minimal subunit-based, chemically synthesized, fully antimalarial vaccine.
A highly robust, sensitive, and specific methodology has thus been designed using 20-mer non-overlapping synthetic peptides in binding assays; this has led to identifying P. falciparum protein HABPs interacting with host cells (Garcia et al. 2006; Rodriguez et al. 2008). Efforts having been focused on conserved HABPs due to P. falciparum proteins’ tremendous genetic polymorphism as a commonly used mechanism for microbes and parasites to evade host immune response.
Every conserved HABP identified so far represents a potential target to be specifically modified to activate an appropriate immune response capable of blocking host–pathogen interactions, i.e. to produce a protective immune response against P. falciparum challenge in the Aotus model (Cifuentes et al. 2009; Patarroyo et al. 2008a; Patarroyo and Patarroyo 2008). Complete CelTOS and TRSP sequences have been finely mapped using HeLa and HepG2 cells, respectively, for identifying specific regions involved in binding to host cells. N-terminal peptides 34451 (21NVLCFRGNNGHNSSSSLYNG40) and 34452 (41SQFIEQLNNSFTSAFLESQSY 60), and C-terminal peptide 34458 (161IWNYNSPDVSESEESLSDDF180) have been identified as HABPs in CelTOS while only TSR domain peptide 36075 (41SDVRYNKSFINNRLLNEHAH60) has been identified as a HABP in TRSP (Fig. 1a, b). Interestingly, two consecutive HABPs were found: 34451 and 34452. An overlapping peptide involving sequences from both these HABPs may also bind to HeLa cells (Lopez et al. 2001; Patarroyo et al. 2008b).
Polymorphism analysis of regions where HABPs were located identified ten non-synonymous substitutions. Four changes within these (shown in bold) were located in CelTOS semi-conserved HABP 34458 (161IWNYNSPDVSESEESLSDDF180) (Fig. 2b). HABP 34451 had just one N-terminal residue substitution regarding the Dd2 strain and the other strains assessed here. By contrast, no changes were observed in the TRSP nucleotide and protein sequence when the 3D7 reference strain was compared with the FCB-2 (Colombia), PAS-2 (unknown origin), and FVO (Vietnam) strains (Fig. 2b). Sequence conservation in these potential vaccine antigens is thus an important issue in blocking P. falciparum parasites’ exquisite immune evasion pathways (Patarroyo and Patarroyo 2008; Hisaeda et al. 2005).
CelTOS and TRSP protein HABPs’ specific binding was determined after HepG2 (for TRSP) and HeLa (CelTOS) cells had been pre-treated with HI, HII, CAC, and CABC to assess the nature of their receptors. CelTOS HABP 34451 and 34458 binding was found to be moderately sensitive to HeLa cell treatment with CABC, CABC activity being mainly directed towards dermatan sulfate and chondroitin-6-, -4-, and -4,6- sulfate (Pradel et al. 2002). Interestingly, it has been reported that CABC has great potential for inhibiting recombinant CSP binding to Kupffer cells, thereby indicating a CSP chondroitin sulfate-dependent interaction during cell traversal (Pradel et al. 2002). The results thus suggest CelTOS HABP interaction with chondroitin sulfate-containing receptors on HeLa cell surface, which could be closely related to dermatan sulfate-like molecules (Fig. 1e); however, additional assays should be performed to confirm such interaction.
Several studies have reported that the TSR domain is involved in protein–protein interactions (Tucker 2004) and that it can also be found in different sporozoite proteins such as CSP (Suarez et al. 2001), TRAP (Lopez et al. 2001), P. falciparum secreted protein with altered thrombospondin repeat (PfSPATR) (Curtidor et al. 2008b) and Plasmodium thrombospondin-related apical merozoite protein (PTRAMP) (Thompson et al. 2004). Studies using the well-described methodology for identifying specific HepG2 cell binding regions have reported that HABPs 3287 and 3289 were found in the TRAP TRS domain (Lopez et al. 2001; Patarroyo et al. 2008b), having poor amino acid sequence similarity with TRSP 36075. However, HABP 36075 binding was slightly sensitive to HI treatment (which cleaves heparin-like oligosaccharides into heparan sulfate, exhibiting a high degree of sulfation and epimerization to iduronic acid) (Fig. 1e); this is similar behavior to that displayed in TRAP heparin sulfate-dependent binding to HepG2 cells. A 1.1 Å RMSD region was observed between TRSP S8-N11 amino acids and TRAP 3289 G8-T11 when 3D structure was superimposed on TRSP HABP 36075 3D structure, suggesting similar structural characteristics (Fig. 3c). A 1.69 Å RMSD was observed when P. falciparum TRAP TSR 3D structure (PDB 2BBX) was superimposed on conserved HABP 36075 structure (determined by NMR) in the corresponding region (data not shown).
Previous studies have reported a 1.55 Å RMSD when TRS protein domain 3D structure was superimposed on conserved HABP 3289 structure in the region corresponding to distorted type III β-turn structures in both molecules (Patarroyo et al. 2008b). However, cross-competition assays revealed that, despite similar structural characteristics and binding to heparan sulfate proteoglycans, HABP 3289 (which also binds specifically to HepG2 cells) did not compete for HABP 36075 binding sites on HepG2 cells (data not shown). This suggested that even though there was structural resemblance between proteins from the same family, both HABPs were involved in different invasion pathways, recognizing subtle receptor differences, as has been widely documented for other molecules (Mayer et al. 2004).
On the other hand, it has been reported that the PfTRAP TSR domain contains a heparin-binding site located in the N-terminal half of the structure (Tossavainen et al. 2006) and that conserved tryptophans (WDEW) together with stacked arginines (RSRKRE) are involved in domain folding (Tossavainen et al. 2006). Interestingly, the TRSP 36076 peptide, whose sequence includes conserved tryptophans (64WSEW67), but not arginines, and is homologous to the TRAP TSR domain N-terminal portion, does not have cell specific binding. Raw data analysis has also indicated that the peptide–cell interaction was non-specific (data not shown). The foregoing shows how minimal changes in sequence can dramatically affect function, this being the basis for our approach.
Previous data have shown that knowledge regarding HABP structure and that of their modified peptides has led to correlating immunological activity with protection against experimental malarial challenge in Aotus spp monkeys. A strong immunogenicity-structure association has been widely reported for regions derived from several P. falciparum merozoite-associated proteins (Reyes et al. 2007) and more recently for the sporozoite-related liver stage antigen-1 (LSA-1), the sporozoite and liver stage antigen (SALSA), CSP and TRAP (Patarroyo et al. 2008b; Bermudez et al. 2008; Cifuentes et al. 2009). This evidence has led to determining CelTOS and TRSP HABP structural content by CD along with NMR for HABP 36075. HABPs 34451, 34452, and 36075 mainly contain α-helical elements in their secondary structures, having two minima at 208 and 220 nm (Fig. 3a). HABP 36075 NMR studies have confirmed the presence of α-helical structures between residues N11-H20 and helical tendency between K7-I10 (Fig. 3b, c). A displacement has been observed in HABP 34458 spectrum (200 nm minima), indicating the presence of some other structural features, such as random coil elements. These results agreed with deconvolution analysis using CONTINLL, SELCON and CDSSTR software, revealing 40% α-helical features for HABP 34458 compared with >95% α-helical elements for HABPs 34451, 34452, and 36075 (Fig. 3a). Such results are relevant since previous studies have shown that specific structural modifications made on α-helical HABPs have been able to induce protective immune responses in Aotus monkeys when immunized with these modified merozoite HABPs (Patarroyo and Patarroyo 2008; Patarroyo et al. 2008a).
Conserved HABPs derived from the relevant proteins observed in P. falciparum merozoite and sporozoite stages are poorly immunogenic and do not induce protection. These HABPs have been used as templates for designing analog peptides to break this immunologic code of silence by substituting critical binding amino acids (determined by glycine analog scanning) for others having the same mass but different polarity, according to previously described physicochemical and biological principles (Patarroyo et al. 2008a). Such detailed changes lead to structural modifications in these analog peptides, thereby allowing a better fit into immune systems molecules and thus improving their immunological characteristics (Patarroyo et al. 2008a).
CelTOS-derived analog peptides 38138 (34451) and 38140 (34458) and TRSP-derived analog 38148 (36075) were thus designed and inoculated in Aotus monkeys to determine their immunogenic properties. Immunofluorescence assays showed that antibodies against modified peptides were able to recognize CelTOS protein as small intracytoplasmic dots, suggesting a micronemal pattern (Fig. 4a red) and TRSP showed a bilobed pattern (Fig. 4a iii, red), suggesting its location in sporozoite rhoptries. Sera from the same Aotus monkeys immunized with these CelTOS- or TRSP-derived modified HABPs have specifically recognized both ~17 kDa rPfCelTOS and ~12 kDa rPfTRSP proteins by WB. Therefore, specific modifications made to HABPs from the different proteins assessed here were able to induce antibodies in the Aotus spp. experimental model which recognized recombinant protein by ELISA and WB and native protein in sporozoites using immunofluorescence. These results stress the importance of complete structural and immunologic analysis for all P. falciparum HABPs for developing multi-epitope, multistage, minimal subunit-based, chemically synthesized vaccines to ensure obtaining complete protection against malaria (Patarroyo et al. 2008a; Patarroyo and Patarroyo 2008). Unfortunately, it was only discovered that native peptide 34458 had genetic polymorphism when polymorphism studies had been completed (as described above), thereby allowing this HABP to be classified as variable according to our strict conservation definition standards and also rule out the possibility of using 38140 modified analog as a component of a minimal subunit-based, synthetic antimalarial vaccine.
This study has thus determined the profile for all CelTOS and TRSP amino acid sequence-derived peptides binding to HeLa and HepG2 cells, respectively, leading to conserved HABPs mainly displaying α-helical features being identified. These HABPs also established high-affinity interactions with heparan-like or dermatan sulfate-containing host-cell surface receptors which have been reported to play an important role in recognition by P. falciparum sporozoite molecules during hepatic cell traversal and invasion. CelTOS and TRSP HABP-derived modified peptides have been rendered highly immunogenic in the Aotus model; they have been able to recognize the recombinant protein in both its native and denatured conformation. Taken as a whole, these results support including these modified CelTOS and TRSP HABPs when designing vaccine components for a multi-stage, multi-epitope, minimal subunit-based, fully-protective antimalarial vaccine, as part of a logical and rational methodology for vaccine development.
References
Akdi RR, Sharma A, Malhotra P, Sharma A (2008) Role of Plasmodium falciparum thrombospondin-related anonymous protein in host-cell interactions. Malar J 7:63
Attie A, Raines R (1995) Analysis of receptor-ligand interactions. J Chem Educ 72:119–124
Bergmann-Leitner ES, Mease RM, De La Vega P, Savranskaya T, Polhemus M, Ockenhouse C, Angov E (2010) Immunization with pre-erythrocytic antigen CelTOS from Plasmodium falciparum elicits cross-species pro1tection against heterologous challenge with Plasmodium berghei. PLoS ONE 5 (8):e12294. doi:10.1371/journal.pone.0012294
Bermudez A, Vanegas M, Patarroyo ME (2008) Structural and immunological analysis of circumsporozoite protein peptides: a further step in the identification of potential components of a minimal subunit-based, chemically synthesised antimalarial vaccine. Vaccine 26(52):6908–6918
Bongfen SE, Ntsama PM, Offner S, Smith T, Felger I, Tanner M, Alonso P, Nebie I, Romero JF, Silvie O, Torgler R, Corradin G (2009) The N-terminal domain of Plasmodium falciparum circumsporozoite protein represents a target of protective immunity. Vaccine 27(2):328–335
Cifuentes G, Bermudez A, Rodriguez R, Patarroyo MA, Patarroyo ME (2008) Shifting the polarity of some critical residues in malarial peptides’ binding to host cells is a key factor in breaking conserved antigens’ code of silence. Med Chem 4(3):278–292
Cifuentes G, Vanegas M, Martinez NL, Pirajan C, Patarroyo ME (2009) Structural characteristics of immunogenic liver-stage antigens derived from P. falciparum malarial proteins. Biochem Biophys Res Commun 384(4):455–460
Combet C, Blanchet C, Geourjon C, Deleage G (2000) NPS@: network protein sequence analysis. Trends Biochem Sci 25(3):147–150
Cowman AF, Crabb BS (2006) Invasion of red blood cells by malaria parasites. Cell 124(4):755–766. doi:S0092-8674(06)00181-4[pii]10.1016/j.cell.2006.02.006
Cowman AF, Baldi DL, Duraisingh M, Healer J, Mills KE, O’Donnell RA, Thompson J, Triglia T, Wickham ME, Crabb BS (2002) Functional analysis of Plasmodium falciparum merozoite antigens: implications for erythrocyte invasion and vaccine development. Philos Trans R Soc Lond B Biol Sci 357(1417):25–33. doi:10.1098/rstb.2001.1010
Curtidor H, Torres MH, Alba MP, Patarroyo ME (2007) Structural modifications to a high-activity binding peptide located within the PfEMP1 NTS domain induce protection against P. falciparum malaria in Aotus monkeys. Biol Chem 388(1):25–36. doi:10.1515/BC.2007.003
Curtidor H, Arevalo G, Vanegas M, Vizcaino C, Patarroyo MA, Forero M, Patarroyo ME (2008a) Characterization of Plasmodium falciparum integral membrane protein Pf25-IMP and identification of its red blood cell binding sequences inhibiting merozoite invasion in vitro. Protein Sci 17(9):1494–1504. doi:ps.036251.108[pii]10.1110/ps.036251.108
Curtidor H, Garcia J, Vanegas M, Puentes F, Forero M, Patarroyo ME (2008b) Identification of peptides with high red blood cell and hepatocyte binding activity in the Plasmodium falciparum multi-stage invasion proteins: PfSPATR and MCP-1. Biochimie 90(11–12):1750–1759. doi:10.1016/j.biochi.2008.08.003
Florens L, Washburn MP, Raine JD, Anthony RM, Grainger M, Haynes JD, Moch JK, Muster N, Sacci JB, Tabb DL, Witney AA, Wolters D, Wu Y, Gardner MJ, Holder AA, Sinden RE, Yates JR, Carucci DJ (2002) A proteomic view of the Plasmodium falciparum life cycle. Nature 419(6906):520–526
Garcia JE, Puentes A, Patarroyo ME (2006) Developmental biology of sporozoite–host interactions in Plasmodium falciparum malaria: implications for vaccine design. Clin Microbiol Rev 19(4):686–707
Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419(6906):498–511
Havel TF, Wuthrich K (1985) An evaluation of the combined use of nuclear magnetic resonance and distance geometry for the determination of protein conformations in solution. J Mol Biol 182(2):281–294
Hay SI, Okiro EA, Gething PW, Patil AP, Tatem AJ, Guerra CA, Snow RW (2010) Estimating the global clinical burden of Plasmodium falciparum malaria in 2007. PLoS Med 7 (6):e1000290. doi:10.1371/journal.pmed.1000290
Hisaeda H, Yasutomo K, Himeno K (2005) Malaria: immune evasion by parasites. Int J Biochem Cell Biol 37(4):700–706. doi:10.1016/j.biocel.2004.10.009
Houghten RA (1985) General method for the rapid solid-phase synthesis of large numbers of peptides: specificity of antigen–antibody interaction at the level of individual amino acids. Proc Natl Acad Sci USA 82(15):5131–5135
Ishino T, Yano K, Chinzei Y, Yuda M (2004) Cell-passage activity is required for the malarial parasite to cross the liver sinusoidal cell layer. PLoS Biol 2(1):E4
Ishino T, Chinzei Y, Yuda M (2005) A Plasmodium sporozoite protein with a membrane attack complex domain is required for breaching the liver sinusoidal cell layer prior to hepatocyte infection. Cell Microbiol 7(2):199–208
Kaiser K, Camargo N, Coppens I, Morrisey JM, Vaidya AB, Kappe SH (2004a) A member of a conserved Plasmodium protein family with membrane-attack complex/perforin (MACPF)-like domains localizes to the micronemes of sporozoites. Mol Biochem Parasitol 133(1):15–26
Kaiser K, Matuschewski K, Camargo N, Ross J, Kappe SH (2004b) Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol Microbiol 51(5):1221–1232
Kappe SH, Buscaglia CA, Nussenzweig V (2004) Plasmodium sporozoite molecular cell biology. Annu Rev Cell Dev Biol 20:29–59
Kariu T, Ishino T, Yano K, Chinzei Y, Yuda M (2006) CelTOS, a novel malarial protein that mediates transmission to mosquito and vertebrate hosts. Mol Microbiol 59(5):1369–1379
Khusmith S, Charoenvit Y, Kumar S, Sedegah M, Beaudoin RL, Hoffman SL (1991) Protection against malaria by vaccination with sporozoite surface protein 2 plus CS protein. Science 252(5006):715–718
Kumar KA, Sano G, Boscardin S, Nussenzweig RS, Nussenzweig MC, Zavala F, Nussenzweig V (2006) The circumsporozoite protein is an immunodominant protective antigen in irradiated sporozoites. Nature 444(7121):937–940. doi:nature05361[pii]10.1038/nature05361
Labaied M, Camargo N, Kappe SH (2007) Depletion of the Plasmodium berghei thrombospondin-related sporozoite protein reveals a role in host cell entry by sporozoites. Mol Biochem Parasitol 153(2):158–166
Lambros C, Vanderberg JP (1979) Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol 65(3):418–420
Lopez R, Curtidor H, Urquiza M, Garcia J, Puentes A, Suarez J, Ocampo M, Vera R, Rodriguez LE, Castillo F, Cifuentes G, Patarroyo ME (2001) Plasmodium falciparum: binding studies of peptide derived from the sporozoite surface protein 2 to Hep G2 cells. J Pept Res 58(4):285–292
Mayer DC, Mu JB, Kaneko O, Duan J, Su XZ, Miller LH (2004) Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA-181 alters its receptor specificity. Proc Natl Acad Sci USA 101(8):2518–2523
Merrifield RB (1963) Solid phase peptide synthesis. I. The synthesis of a tetrapeptide. J Am Chem Soc 85:2149–2154
Mongui A, Angel DI, Moreno-Perez DA, Villarreal-Gonzalez S, Almonacid H, Vanegas M, Patarroyo MA (2010) Identification and characterization of the Plasmodium vivax thrombospondin-related apical merozoite protein. Malar J 9:283. doi:10.1186/1475-2875-9-283
Patarroyo ME, Patarroyo MA (2008) Emerging rules for subunit-based, multiantigenic, multistage chemically synthesized vaccines. Acc Chem Res 41(3):377–386
Patarroyo ME, Cifuentes G, Bermudez A, Patarroyo MA (2008a) Strategies for developing multi-epitope, subunit-based, chemically synthesized anti-malarial vaccines. J Cell Mol Med 12(5B):1915–1935
Patarroyo ME, Cifuentes G, Rodriguez R (2008b) Structural characterisation of sporozoite components for a multistage, multi-epitope, anti-malarial vaccine. Int J Biochem Cell Biol 40(3):543–557
Pradel G, Garapaty S, Frevert U (2002) Proteoglycans mediate malaria sporozoite targeting to the liver. Mol Microbiol 45(3):637–651
Rathore D, Hrstka SC, Sacci JB Jr, De la Vega P, Linhardt RJ, Kumar S, McCutchan TF (2003) Molecular mechanism of host specificity in Plasmodium falciparum infection: role of circumsporozoite protein. J Biol Chem 278(42):40905–40910
Reyes C, Patarroyo ME, Vargas LE, Rodriguez LE, Patarroyo MA (2007) Functional, structural, and immunological compartmentalisation of malaria invasive proteins. Biochem Biophys Res Commun 354(2):363–371
Roccatano D, Colombo G, Fioroni M, Mark AE (2002) Mechanism by which 2, 2, 2-trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: a molecular dynamics study. Proc Natl Acad Sci USA 99(19):12179–12184. doi:10.1073/pnas.182199699
Rodriguez LE, Curtidor H, Urquiza M, Cifuentes G, Reyes C, Patarroyo ME (2008) Intimate molecular interactions of P. falciparum merozoite proteins involved in invasion of red blood cells and their implications for vaccine design. Chem Rev 108(9):3656–3705
Sinnis P, Coppi A (2007) A long and winding road: the Plasmodium sporozoite’s journey in the mammalian host. Parasitol Int 56(3):171–178
Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI (2005) The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434(7030):214–217
Sreerama N, Woody RW (2000) Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem 287(2):252–260
Suarez JE, Urquiza M, Puentes A, Garcia JE, Curtidor H, Ocampo M, Lopez R, Rodriguez LE, Vera R, Cubillos M, Torres MH, Patarroyo ME (2001) Plasmodium falciparum circumsporozoite (CS) protein peptides specifically bind to HepG2 cells. Vaccine 19(31):4487–4495
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24 (8):1596–1599. doi:10.1093/molbev/msm092
Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2 3. doi:10.1002/0471250953.bi0203s00
Thompson J, Cooke RE, Moore S, Anderson LF, Janse CJ, Waters AP (2004) PTRAMP; a conserved Plasmodium thrombospondin-related apical merozoite protein. Mol Biochem Parasitol 134(2):225–232. doi:10.1016/j.molbiopara.2003.12.003
Tossavainen H, Pihlajamaa T, Huttunen TK, Raulo E, Rauvala H, Permi P, Kilpelainen I (2006) The layered fold of the TSR domain of P. falciparum TRAP contains a heparin binding site. Protein Sci 15(7):1760–1768. doi:15/7/1760[pii]10.1110/ps.052068506
Tucker RP (2004) The thrombospondin type 1 repeat superfamily. Int J Biochem Cell Biol 36(6):969–974
Wüthrich K (New York 1986) NMR of proteins and nucleic acids. Wiley
Yuda M, Ishino T (2004) Liver invasion by malarial parasites—how do malarial parasites break through the host barrier? Cell Microbiol 6(12):1119–1125
Acknowledgments
We would like to thank Jason Garry for translating this manuscript.
Conflict of interest
The authors declare no conflict of interest. The authors alone are responsible for the content and writing of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Curtidor, H., Arévalo-Pinzón, G., Bermudez, A. et al. Binding activity, structure, and immunogenicity of synthetic peptides derived from Plasmodium falciparum CelTOS and TRSP proteins. Amino Acids 43, 365–378 (2012). https://doi.org/10.1007/s00726-011-1087-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00726-011-1087-8